Professor James Nathan
Cellular mechanisms of oxygen and metabolite sensing
Affiliation: Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID)
Research Summary
Oxygen-sensing and chromatin modifications
A fundamental requirement for cells is the ability to adapt to local oxygen and nutrient environments. Our goal is to gain novel insights into how oxygen is sensed in cells and can govern cell-fate decisions through epigenetic modifications and transcriptional pathways.
Organisms have adopted several strategies to monitor oxygen abundance and adapt. At a cellular level this involves several oxygen-sensitive enzymes that ‘sense’ oxygen availability and govern cell fates through a dedicated transcriptional pathway, termed the Hypoxia Inducible Factor (HIF) Response, or through oxygen-sensitive enzymes that govern epigenetic marks.
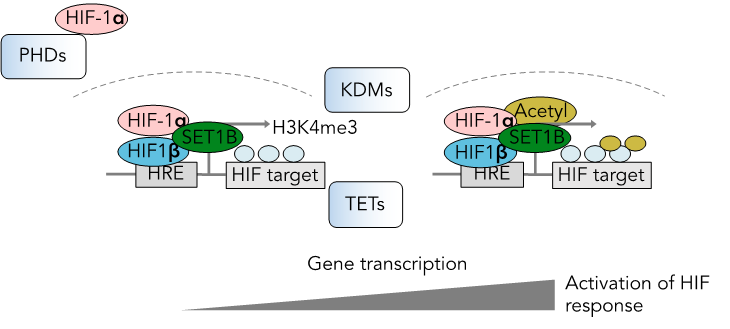
We use a combination of genome-wide mutagenesis screens and metabolic assays to understand how the oxygen-sensitive transcriptional pathways work, and the implications for cell function. A current goal is to understand how chromatin marks can be selectively modified by changes in metabolite or oxygen abundance. This is exemplified by our identification of hypoxic recruitment of the SET1B histone methyltransferase to chromatin, and the regulation of DNA and histone methylation by metabolites.
Regulation of the HIF response at the level of chromatin. PHDs, KDMs and TETs are oxygen and metabolite sensitive.
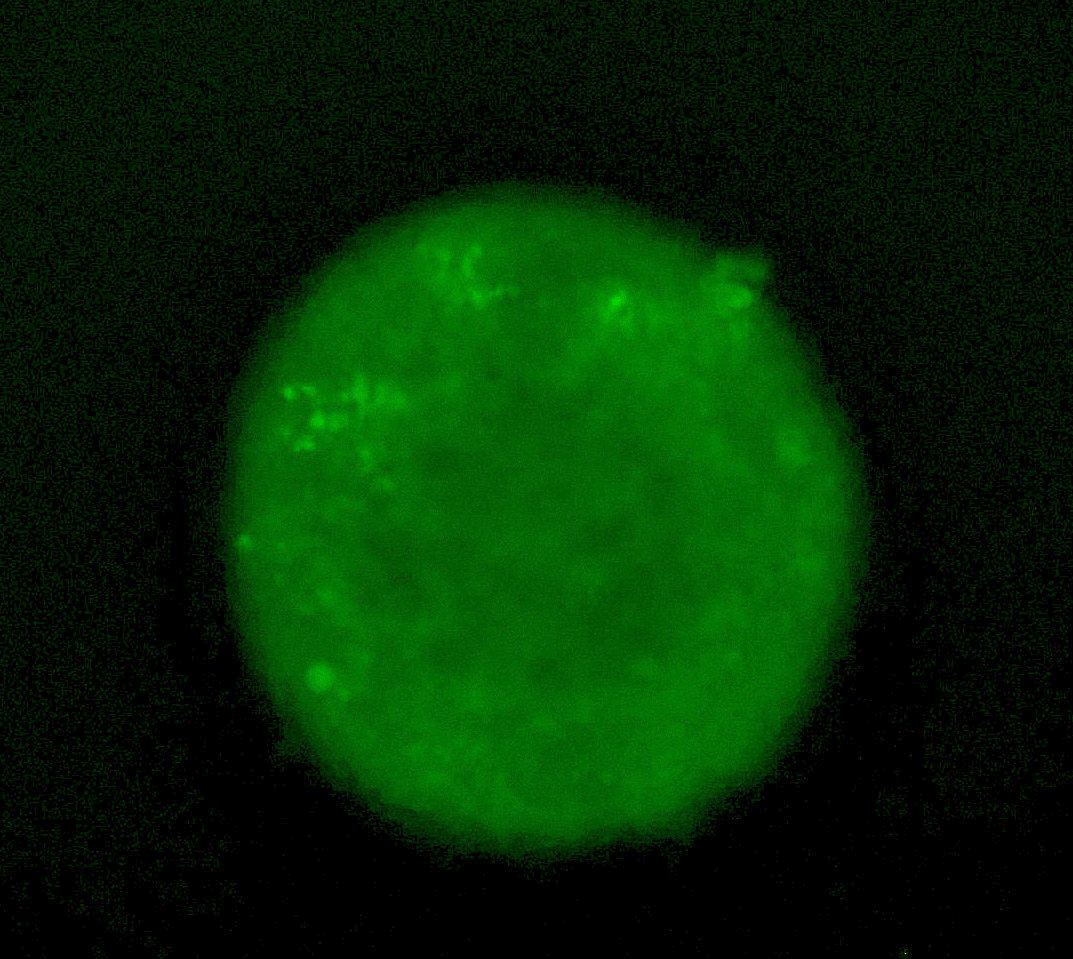
Tumour spheroid showing induction of the HIF response with a fluorescent GFP reporter.