Dr Namshik Han
AI Drug Discovery
Departmental affiliation: Milner Therapeutics Institute and Cambridge Centre for AI in Medicine
Email: nh417@cam.ac.uk
Biography
Han is a computational drug discovery scientist with expertise in machine learning, computational biology and multi-omics. He currently holds the position of Head of Computational Research & AI at Milner Therapeutics Institute, as well as an Associate Faculty position at Cambridge Centre for AI in Medicine, both within the University of Cambridge. Additionally, he serves as an Adjunct Professor at Yonsei University College of Medicine. Han's research focuses on developing and applying specialized Artificial Intelligence technologies to unravel complex multi-modal biomedical datasets, unveiling new disease pathways and mechanisms.
Beyond academia, Han is driven by the opportunity to bridge the gap between academia and industry, particularly in the field of therapeutics and patient care. AI has the potential to revolutionise disease mechanism identification, drug target discovery, and the entire spectrum of patient care, from diagnosis to treatment. He facilitates access to state-of-the-art AI technology for partner organisations within the Milner Consortium, while also developing novel computational methods to fulfil our global mission of identifying innovative therapies through the analysis of big data. In addition, Han played a role in establishing Storm Therapeutics and co-founded two start-ups. The first, KURE.ai Therapeutics in the USA, focuses on the development of immune-oncology NK cell therapy, while the second, CardiaTec Biosciences in the UK, is dedicated to innovative drugs for cardiovascular diseases.
Research Summary
The primary objective of Han's lab at the Milner Therapeutics Institute is to enhance cutting-edge Artificial Intelligence technology for therapeutics research. They strive to develop innovative computational methods that align with the global mission of discovering new and improved therapies through the analysis of large-scale biomedical data. By fostering an interdisciplinary environment, they enable seamless translation and validation of findings from virtual simulations to real-world models, spanning across various therapeutic areas. The core focus of the lab lies in the advancement and application of machine learning, statistical analysis, and mathematical techniques in the fields of multi-omics and drug discovery. They place strong emphasis on harnessing publicly available big data and leveraging purposefully generated experimental data from academic collaborators and pharma/biotech partners. Within the group, they undertake projects that encompass diverse areas, such as:
- Exploring and identifying novel therapeutic targets for a wide range of diseases.
- Identifying opportunities for drug (re)positioning, uncovering alternative applications for existing medications.
- Predicting the efficacy and safety of both new and existing drugs.
- Stratifying patients to enhance the effectiveness of personalized medicine approaches.
Through these endeavours, Han aims to contribute significantly to the progressive environment of the Milner Therapeutics Consortium, facilitating the dynamic translation and validation of research findings. Ultimately, their efforts pave the way for transformative advancements in the field of biomedical research and enable the development of life-changing therapies.
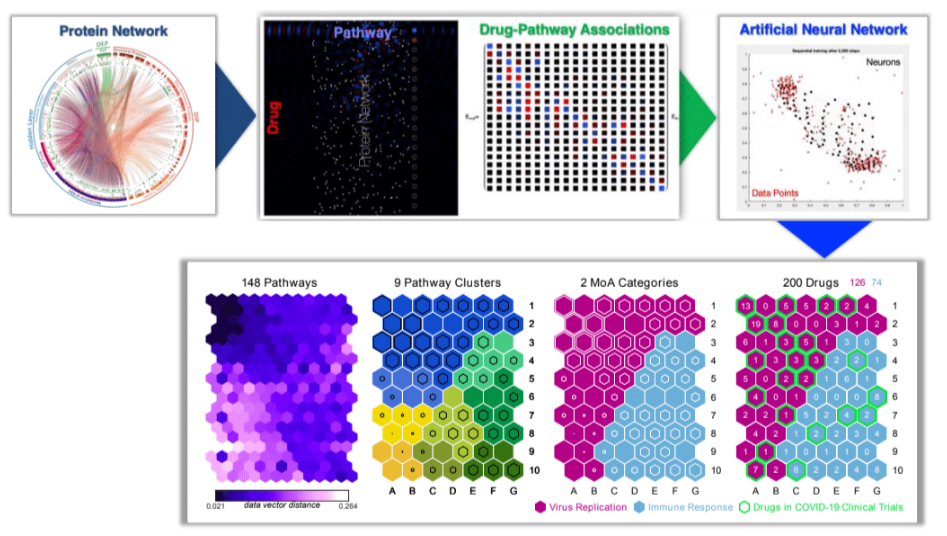
The lab utilized Artificial Neural Networks (ANNs) to predict the Mode of Action (MoA) of drugs. To train the network, they simulated the drugs on the protein-protein interaction network. This approach aimed to capture the interactions between the drugs and specific proteins. By learning from this simulated data, the ANN can recognize patterns and associations between drugs and their MoA. This predictive model has the potential to aid in drug discovery by providing insights into the mechanisms of action of newly identified drugs. Han et al., Science Advances (2021) https://doi.org/10.1126/sciadv.abh3032
CSCI Collaborators
Additional Links
https://www.bio.cam.ac.uk/staff/namshik-han
https://www.milner.cam.ac.uk/machinelearning/