Dr Joo-Hyeon Lee
Stem cells and niches
Email: jhl62@cam.ac.uk
Laboratory: Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre
Departmental Affiliation: Physiology, Development and Neuroscience
Biography
Joo-Hyeon Lee is an Associate Professor in Stem Cell Medicine, a Wellcome Senior Research Fellow at the Wellcome – MRC Cambridge Stem Cell Institute, and an affiliated faculty member at the Department of Physiology, Development, and Neuroscience (PDN), University of Cambridge. Her research interests focus on understanding the mechanisms of stem cell fate regulation. Her lab particularly studies how stem cells sense environmental changes and determine their cell fate, and how niches develop and remodel the local environment during lung homeostasis, regeneration and the early stages of disease progression. Her group develops the state-of-the art technologies, including in vitro 3D human lung organoids and in vivo genetic labelling models, and employs computational analyses of single cell multi-omics and clonal biophysical modelling to map lineage hierarchy and stem-niche interactions at the single cell level.
Joo-Hyeon received a Wellcome Trust Sir Henry Dale Fellowship, an ERC starting grant, a Suh Kyoungbae Foundation Young Investigator award, and a Wellcome Senior Research Fellowship.
Funding
Wellcome Trust, Suh Kyungbae Foundation, BBSRC, AstraZeneca
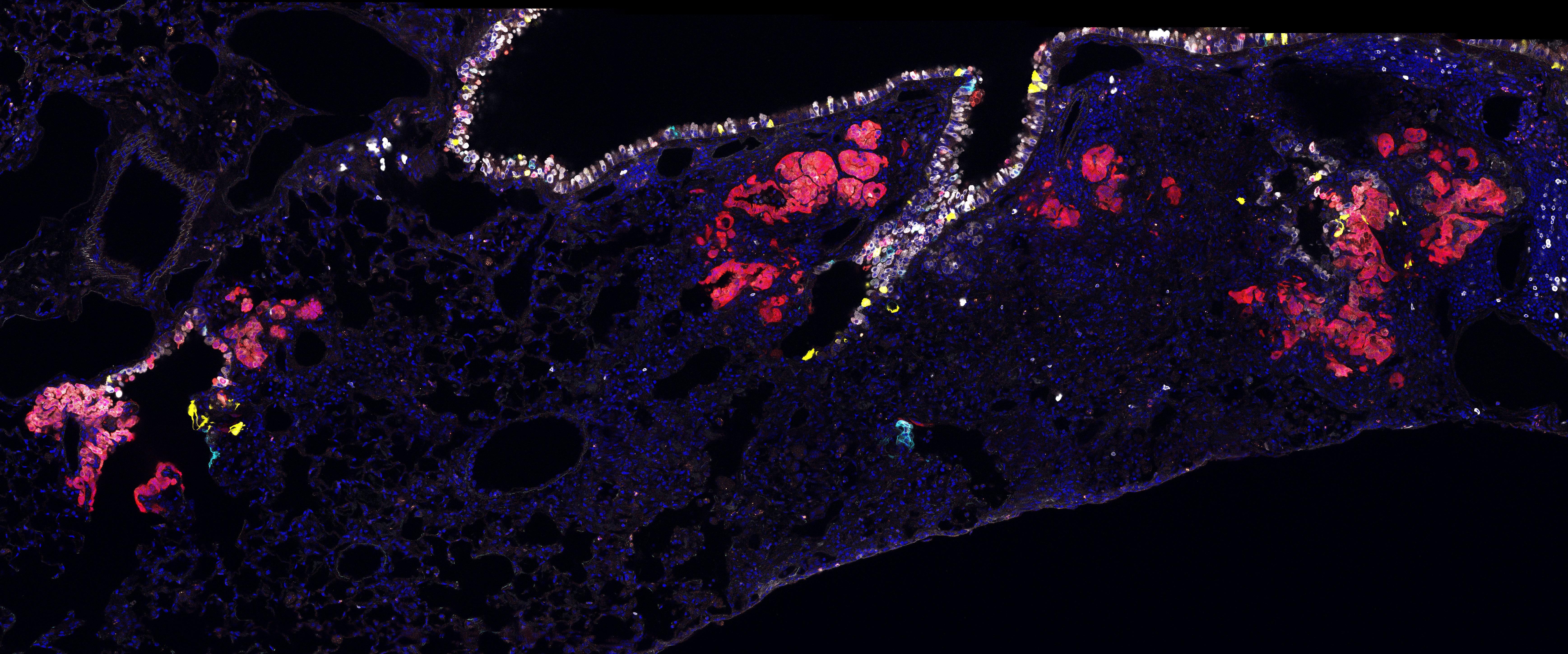
Clonal expansion of lineage labelled progenitor cells in mouse lungs (Image credit: Catherine Dabrowska)
Research
What are the regulatory mechanisms that control homeostatic turnover, and how do their perturbation contribute to disease progression? The lung is a very slow cycling organ that is composed of diverse epithelial and stromal cell types, but has capacity to rapidly regenerate new cells after injury. Lee group is trying to understand how stem cells respond to different signals from their local environment and orchestrate the changes in chromatin, transcription, translation, and cellular dynamics in homeostasis and injury repair. We investigate the regulatory networks that need to be turned on and off at the right time and place for stem cells to become activated and generate specialised cell types during regeneration. We are also interested in defining cellular heterogeneity and plasticity during this process. Elucidating the normal process of lung dynamics will provide us a foundation to understand lung diseases and cancer. We couple ex vivo 3D organoid cultures of human and mouse lungs with genetic tools, in vivo transgenic mouse models with lineage tracing techniques, quantitative mathematic modelling of clonal dynamics, and bioinformatics at the single cell level.
Group Members
Frances England, JaeHak Bang, Erik Cardoso, Jakub Chundziak, Christopher Lambert, Minn-E Ng, Bumsoo Kim, Keat Ying Chan
Opportunities:
If you are interested in working with us, please contact us at jhl62@cam.ac.uk.
Plain English
Our interest is focused on understanding how the diverse epithelial and stromal cells are maintained for a lifetime and regenerated following injury in the lungs. We study the regulatory mechanisms that balance stem cell maintenance and differentiation, and how their perturbation contribute to disease progression.
Key Publications
-
Lee W*, Lee S*, Yoon JK*, Lee D, Kim Y, Han YB, Kim R, Moon, S, Park YJ, Park K, Cha B, Choi J, Kim J, Ha NY, Kim K, Cho S, Cho NH, Desai T.J., Chung JH, Lee JH†, Kim JI† (2023). A single cell atlas of in vitro multi-systems uncovers in vivo lineage trajectory and cell state in the human lung. Experimental & Molecular Medicine. In press. *Equal contribution; †Corresponding authors.
-
Varankar S, Cardoso E.C., Lee JH (2022). Ex situ-armus: Experimental models for combating respiratory dysfunction. Current Opinion in Genetics & Development. 2022 Aug;75:101946.
-
Jeon HY*, Choi J*, Kraaier L, Kim YH, Eisenbarth D, Yi K, Kang JG, Kim JW, Shim HS, Lee JH† and Lim DS† (2022). Airway secretory cell fate conversion via YAP-mTORC1-dependent essential amino acid metabolism. EMBO J. 2022 Mar 14;e109365. *Equal contribution; †Corresponding authors.
-
Choi J*, Jang YJ*, Dabrowska C, Iich E, Evans K.V, Hall H, Janes S.M, Simons B.D, Koo BK, Kim J, Lee JH† (2021). Release of Notch signalling coordinated by inflammation confers cross-compartment differentiation plasticity during alveolar regeneration. Nature Cell Biology. 2021 Sep;23(9):953-966.
-
Liu Y, Dabrowska C, Mavousian A, Strauss B, Meng F, Mazzaglia C, Ouaras K, Macintosh C, Terentijev E, Lee JH† and Huang Y Y S† (2021) Bio-assembling Macro-Scale, Lumenized Airway Tubes of Defined Shape via Multi-Organoid Patterning and Fusion. Advanced Science. 8(9):2003332. PMID: 33977046. †Corresponding authors.
-
Evans K.V. and Lee JH (2020). Alveolar Wars: The rise of in vitro models to understand human lung alveolar maintenance, regeneration, and disease. Stem Cells Translational Medicine. 2020 Aug;9(8):867-881.
-
Youk J*, Kim T*, Evans KV*, Jeong YI*, Hur Y, Hong SP, Kim JH, Yi K, Kim SY, Na KJ, Bleazard T, Kim HM, Fellows M, Mahbubani KT, Saeb-Parsy K, Kim SY, Kim YT†, Koh GY†, Choi BS†, Ju YS† and Lee JH† (2020) Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell. 27(6):905-919.e10. PMCID: PMC7577700 *Equal contribution. †Corresponding authors.
-
Choi J, Park JE, Tsagkogeorga G, Yanagita M, Koo BK, Han N, Lee JH (2020). Inflammatory signals induce AT2 cell-derived Damage-Associated Transient Progenitors that mediate alveolar regeneration. Cell Stem Cell. 27(3):366-382.e7. PMCID: PMC7487779
-
Ombrato L, Nolan E, Kuerlac I, Mavousian A, Bridgeman V, Heinze I, Chakravarty P, Horswell S, Gonzales-Gualda E, Matacchione G, Weston A, Kirkpatrick J, Husain E, Speirs V, Collinson L, Ori A, Lee JH† and Malanchi I† (2019). Metastatic niche labelling reveals tissue parenchyma stem cell features. Nature. 572 (7771), 603-608. PMCID: PMC6797499 †Corresponding authors
-
Lee JH†, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner DA, Wu K, Paschini M, Bhang DH, Jacks T, Regev A, Kim CF† (2017). Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell. 2017 Sep 7;170(6):1149-1163.e12. PMCID: PMC5607351 †corresponding authors
-
Lee JH, Bhang DH, Beede A, Huang TL, Stripp B, Bloch KD, Wagers AJ, Tseng YH, Ryeom S, Kim CF (2014) Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1- Thrombospondin-1 axis. Cell. 156(3):440-55. PMCID:PMC3951122



