Submitted by Laura Puhl on Mon, 08/01/2024 - 13:25
A new collaborative paper from the Göttgens, Kranc and O’Carroll groups reports the first model of homeostatic in vivo tissue dynamics that is anchored in single cell genomics data in the Cell Stem Cell publication out last week (5 January).
In time series measurements extending over 9 months, the Kranc Group (Institute of Cancer Research, London) and the Göttgens Group performed single cell transcriptomics of over 130,000 cells to track the fates of blood stem cells in normal mouse bone marrow.
Tracking the cells was made possible by using a new transgenic mouse model generated by the O’Carroll lab (Edinburgh) and validated by the O’Carroll and Kranc labs. The Göttgens team went on to use the novel time-series dataset to generate new time and single-cell resolved computational models capturing the tissue dynamics of murine bone marrow haematopoiesis. This allowed the coupling of cascading single-cell expression patterns with dynamic changes in differentiation and growth speeds.
The resulting explicit linkage between molecular states and cellular behaviour revealed widely varying self-renewal and differentiation properties across distinct lineages. Moreover, transplanted stem cells showed strong acceleration of specific differentiation steps, thus illustrating how the model quantifies the impact of perturbations.
Bertie Göttgens says, “I am so pleased to see this tremendous team effort come to fruition. This study required close collaboration between groups and across disciplines, and already forms the basis of much of our ongoing and planned research.”
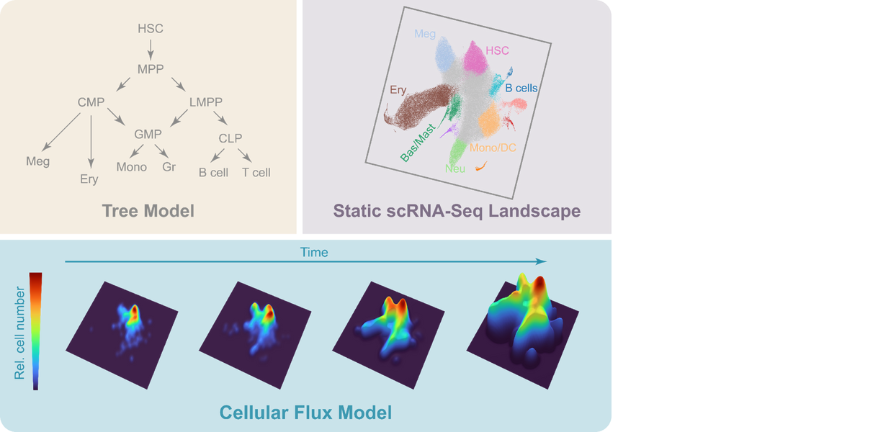
A time and single-cell resolved model of haematopoiesis: The classical population-based differentiation tree model (top left) facilitated the derivation of quantitative tissue flux models (e.g. Busch et al, Nature 2015), but is not compatible with more recent singe cell ‘omics differentiation landscapes (top right). Persistent HSC labelling combined with time-series scRNA-Seq enabled the generation of a new quantitative and predictive Cellular Flux Model (lower part), which captures in vivo cell behaviour in real-time and connects it with gene expression patterns. This broadly applicable approach will empower the single cell genomics community to dissect tissue scale dynamics in both health and disease.
The ever-increasing throughput of modern single cell ‘omics methods means that datasets encapsulating the cellular complexity of entire organ systems can now be generated, including the many intermediate cell states present in continuously replenishing organs such as blood, intestine, or skin. However, ‘omics protocols deliver snapshot measurements, because cells need to be destroyed to characterize their molecular states. As a result, it’s impossible to determine from those measurements how long it takes for a given cell to complete a differentiation process, nor at which precise stages cell numbers expand or contract. Changes in tissue dynamics underlie many major diseases, such as ageing associated defects in tissue maintenance and regeneration as well as clonal stem cell competition during early premalignant growth, but in order to quantify such defects, researchers must first define tissue dynamics in the unperturbed healthy state.
To deliver this critical information for the blood system, previous studies had employed flow cytometric approaches. A key early result of the single cell genomics revolution was that flow cytometry-derived hierarchies are unable to capture the full complexity of blood stem/progenitor cell differentiation because flow cytometric populations are neither functionally nor molecularly homogeneous. Moreover, their differentiation status was commonly defined by potency assays, which reveal what a given cell can do when placed out of its natural context, but this may be different from what the same cell would end up doing if left alone.
For the first time, the new research from Göttgens, Kranc and O’Carroll provides a strategy to create predictive dynamic models of a regenerative organ in vivo, with the capacity to model the response to subtle changes, at single cell resolution and over extended timeframes. As this approach is generally applicable to understanding tissue scale dynamics at high resolution in other human organs, its application in blood research and beyond will not only illuminate normal organ function, but also deliver the critical reference data to quantify how perturbation of tissue dynamics lies at the heart of a multitude of diseases that represent major health challenges.