Dr Jyoti Nangalia
Somatic mutagenesis and clonal evolution in blood
Email: jn218@cam.ac.uk
Laboratory: Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre
Departmental Affiliation: Haematology;
Biography
Jyoti Nangalia studied Medicine at Cambridge University and subsequently trained as a Haematologist. She undertook a PhD at the Cambridge Institute of Medical Research where she developed a passion for genomics. During her PhD, she discovered CALR mutations in the vast majority of patients with JAK2 unmutated myeloproliferative neoplasms (MPN). Testing for CALR mutations in clinical practice is now routine. Utilising cancer genetics to support clinical decision-making is a critical application of genetic sequencing technologies, and following her PhD, she worked on the development of an accurate online personalised predictor of prognosis for patients with blood cancer by integrating clinical and genomic parameters. Her research group focuses on using sequencing technologies to understand exactly when blood cancers originate in patients during their lifetime, their clonal trajectories and the landscape of selection on driver mutations in blood. Her team studies somatic mutagenesis and epigenetic readouts to understand the changes in blood over lifespan, from development to ageing, pre malignancy to cancer. She is currently a Principal Investigator at the Wellcome-MRC Cambridge Stem Cell Institute and a Cancer Research UK Clinician Scientist at the Wellcome Sanger Institute with a group shared between both institutes. She is a Consultant Haematologist and treats patients with MPN. She is a member of the MPN clinical study subgroup of the National Cancer Research Institute and has contributed to the writing of national guidelines for MPN in the UK and analysis of MPN clinical trials.
Funding
CRUK, Rosetrees, Alborada, MPNRF
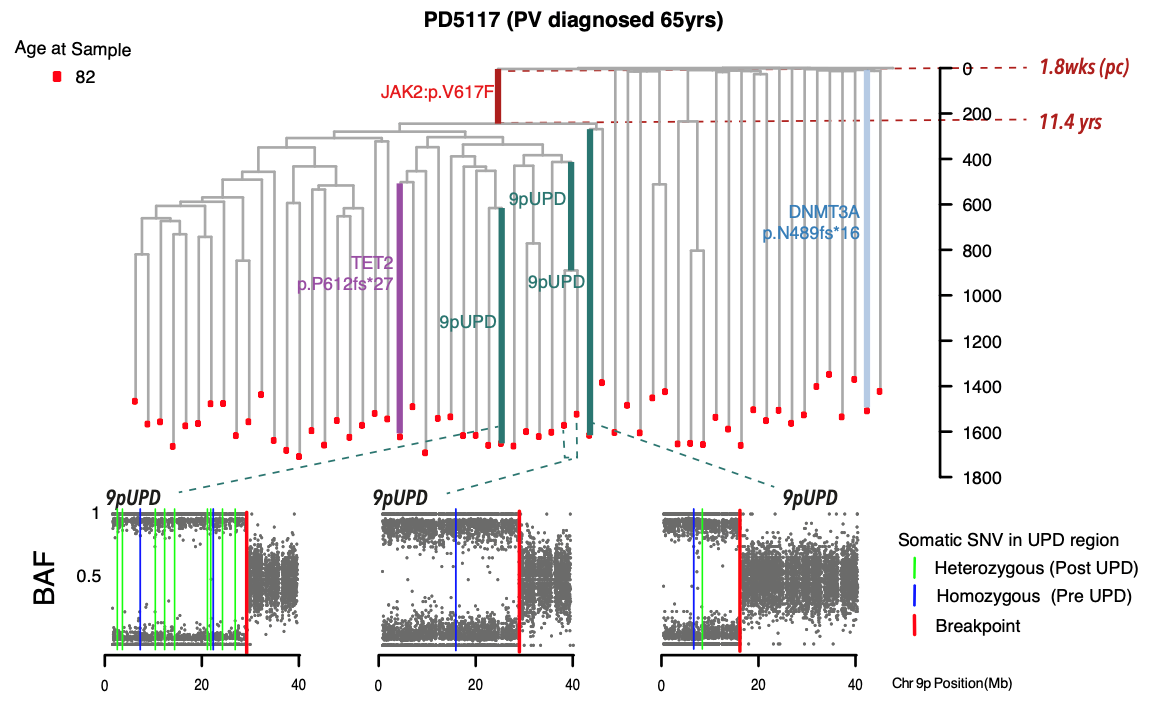
A family tree of blood production in a patient who developed an MPN at age 65, showing the acquisition of the disease causing JAK2 mutation over 50 years before disease presentation, changing the current paradigm for the evolution of this blood cancer. We use single cell derived clonal whole genome sequencing to trace the timing of blood cancer, and track its evolution over the life of individuals (image generated by Nick Williams).
Research
The Nangalia group is currently investigating:
- Clonal trajectories to blood cancers. they are exploring when different types of blood cancers commence their growth during the lifetime of patients. Studying the somatic mutation phylogenies, selection landscape and clonal evolutionary dynamics across haematological diseases and cancers will better inform effective early intervention strategies.
- Methylation dynamics in normal tissues: the lab is studying how the methylome changes over the human lifespan in healthy and diseased tissues using a phylogenetic approach.
- Personalised cancer modelling: the group continues to enhance our blood cancer prognostic tools and build further predictive models for other cancers.
|
Interview following our late breaking abstract at the American Society of Haematology 2020: |
|
Introduction to myeloproliferative neoplasms for the general audience: |
Introduction to the MPN personalised predictive model: http://quadia.webtvframework.com/quadia/_app/player_viewing/?id=3351986
Personalised predictive model for patients with myeloproliferative neoplasms: https://www.sanger.ac.uk/science/tools/progmod/progmod/
Plain English
In our lab we study human blood samples across lifespan from development to ageing and precancer to cancer. We also have a particular interest in myeloproliferative neoplasms, a chronic form of blood cancer.
Key Publications
-
Machado HE*, Obro NF*, Williams N*, … Kent DG*, Nangalia J*, Warren AJ* Convergent somatic evolution commences in utero in a germline ribosomopathy. Nature Comms Aug 2023
-
Williams N, Lee J, Mitchell E, …Green AR*, Campbell PJ*, Nangalia J*. Life histories of myeloproliferative neoplasms inferred through phylogenies. Nature Feb 2022
-
Mitchell E, Spencer Chapman M, Williams N, …Nangalia J*, Laurenti E*, Campbell PJ*. Clonal dynamics of haematopoiesis across the human lifespan. Nature Jun 2022
-
Grinfeld J, Nangalia J, Baxter EJ, Wedge DC, Angelopoulos N, Cantrill R, Godfrey AL, Papaemmanuil E, Gundem G, MacLean C, Cook J, O'Neil L, O'Meara S, Teague JW, Butler AP, Massie CE, Williams N, Nice FL, Andersen CL, Hasselbalch HC, Guglielmelli P, McMullin MF, Vannucchi AM, Harrison CN, Gerstung M, Green AR, Campbell PJ. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. Multicenter Study. N Engl J Med. 2018 Oct 11;379(15):1416-1430. PMCID: PMC7030948
-
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020 Feb;578(7793):82-93. PMCID: PMC7025898
-
Nangalia J, Campbell PJ. Genome Sequencing during a Patient's Journey through Cancer. Review. N Engl J Med. 2019 Nov 28;381(22):2145-2156.
-
McMullin MFF, Mead AJ, Ali S, Cargo C, Chen F, Ewing J, Garg M, Godfrey A, Knapper S, McLornan DP, Nangalia J, Sekhar M, Wadelin F, Harrison CN; A guideline for the management of specific situations in polycythaemia vera and secondary erythrocytosis: A British Society for Haematology Guideline. Br J Haematol. 2019 Jan;184(2):161-175. PMCID: PMC6519221
-
Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, Aziz A, Godfrey AL, Hinton J, Martincorena I, Van Loo P, Jones AV, Guglielmelli P, Tarpey P, Harding HP, Fitzpatrick JD, Goudie CT, Ortmann CA, Loughran SJ, Raine K, Jones DR, Butler AP, Teague JW, O'Meara S, McLaren S, Bianchi M, Silber Y, Dimitropoulou D, Bloxham D, Mudie L, Maddison M, Robinson B, Keohane C, Maclean C, Hill K, Orchard K, Tauro S, Du MQ, Greaves M, Bowen D, Huntly BJP, Harrison CN, Cross NCP, Ron D, Vannucchi AM, Papaemmanuil E, Campbell PJ, Green AR. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013 Dec 19;369(25):2391-2405. PMCID: PMC3966280