Professor Brian Huntly
Leukaemia stem cell biology and leukaemogenesis
Email: bjph2@cam.ac.uk
Laboratory location: Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre
Departmental Affiliation: Haematology
Biography
Prof Brian Huntly is a clinical research scientist who combines running a laboratory group with his practice as a Consultant Haematologist in Addenbrooke’s Hospital. He is also Head of the Department of Haematology at the University of Cambridge. Internationally, he is a member of the European Haematology Association Executive Board and Research Committee and Chair of their Fellowships and Grants Committee. He studied Medicine at Edinburgh, trained in Haematology in Dundee and Cambridge and is a member of the Royal College of Physicians and a Fellow of the Royal College of Pathologists. He studied for his PhD in Cambridge and performed post-doctoral work at Harvard, prior to winning the EHA-José Carreras Young Investigator Award and returning to Cambridge to set up his own research group. The interest of his group is in understanding how normal stem and progenitor cell function is subverted during the step-wise evolution of haematological malignancies, particularly acute myeloid leukaemia (AML) and malignant lymphomas. The group focus particularly on transcriptional and epigenetic alterations and combine functional, genomic and proteomic techniques in mouse and cell line models, as well as human primary tumour tissue, to identify disease mechanisms and potential therapeutic targets. Notable achievements include:
- The first demonstration that chronic and acute myeloid leukaemia may arise in separate stem and progenitor cells (Cancer Cell 2004);
- The discovery (with Tony Kouzarides) of an essential role for BET transcriptional regulators in AML and showed that BET inhibitors provide a novel therapeutic approach (Nature 2011)
- The discovery that malignant lymphomas may initiate in stem and progenitor cells prior to lymphoid commitment (Nature Cell Biology 2017);
- Establishing (with George Vassiliou) that tumour suppression by UTX/KDM6a is mediated by enhancer and chromatin remodeling but that it does not require catalytic activity (Nature Genetics 2018).
Funding
AstraZeneca, Cancer Research UK, European Hematology Association, European Research Council, Kay Kendall Leukaemia Fund, La Caixa Health Foundation, Lady Tata Memorial Trust, Medical Research Council, NIHR Cambridge Biomedical Research Centre, Wellcome Trust
External links
http://www.haem.cam.ac.uk/staff/senior-staff/dr-brian-huntly/
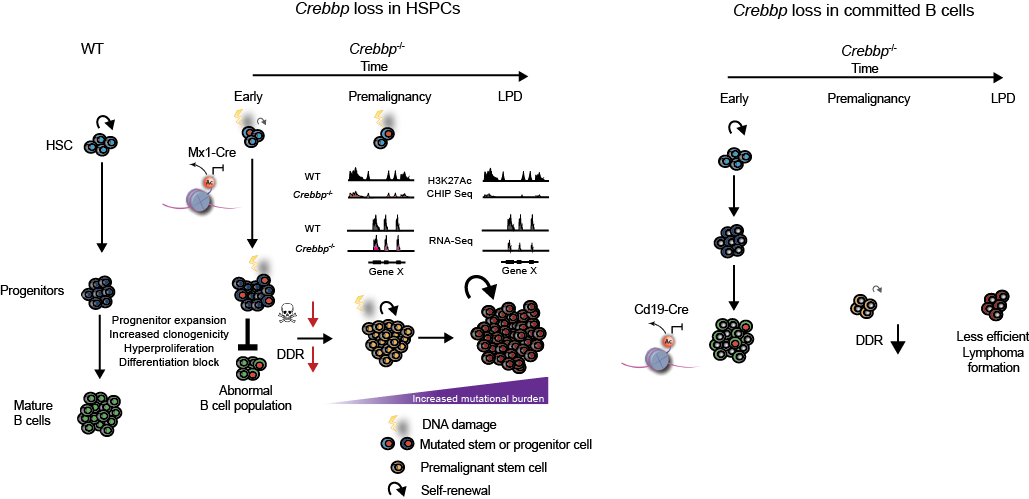
Model of mechanisms and stage specific nature of Crebbp tumour suppression in lymphoid malignancies. Following early loss of Crebbp activity in the HSPC compartment, abnormal epigenetic regulation, post-translational modifications of substrate proteins and altered transcription lead to expansion of lymphoid progenitors, blocked differentiation, increased proliferation and clonogenicity within this expanded progenitor pool. In association with an alteration of the DNA damage response mediated through suboptimal Crebbp acetylation of p53, DNA double strand breaks accumulate and accompanied by a relative decrease in apoptosis, lead to the retention of mutations within B-cell progenitors. We illustrate the epigenetic priming of specific loci at earlier time points, where H3K27ac CHIP-seq and RNA-seq at the critical gene X locus read out in terms of downregulation of gene expression only at later timepoints during lymphoma evolution. We speculate that the molecular aberrations cumulatively acquired interact with this primed epigenetic state and lead to the restoration of self-renewal in this abnormal pre-malignant stem cell population and to the evolution of lymphoma.
Research
Leukaemias have been demonstrated to be wholly dependent upon a small population of so-called cancer stem cells. These cells represent the critical targets for cancer treatment and a greater understanding of their biology and its interface with normal stem cell function is fundamental to improving treatment outcomes.
The focus of the Huntly laboratory is on this interface. They use a combination of techniques in cell lines, animal models and primary human tissue to dissect stem cell function. The aim is to understand how normal stem cell function is corrupted during the evolution and maintenance of cancer and how these processes might be therapeutically targeted to improve the outcome in haematological malignancies. They are examining the role of mutations that occur in and alter the role of haematopoietic stem and progenitors. Many of these mutations alter epigenetic regulation, enhancer function and transcriptional and metabolic programmes and these are all ongoing areas of investigation within the lab.
Much of the lab’s research focuses on lymphoma and acute myeloid leukaemia (AML), a disease with dismal five-year survival rates and urgent clinical need for new treatment options. Therapeutically, their work includes the identification of the Bromodomain and extra terminal (BET) proteins as critical mediators of leukaemia stem cells in AML and the development of an inhibitor of these proteins in a Phase I/II clinical trial in relapsed blood cancers.
Huntly Group photo
Plain English
Our group is interested in the interface between normal and malignant haematopoietic stem cell biology. Comparisons of these systems will allow us to determine how normal regulatory mechanisms are corrupted to generate haematological malignancies. We use experimental model systems and patient samples to answer these questions. This information will provide the basis for targeted therapies to improve the often dismal treatment outcomes for haematological cancers.
Key Publications
-
Lara-Astiaso D, Goñi-Salaverri A, ... Huntly BJP. In vivo screening characterizes chromatin factor functions during normal and malignant hematopoiesis. Nat Genet. 2023 Sep;55(9):1542-1554. PMCID: PMC10484791
-
Agrawal-Singh S, Bagri J, ... Huntly BJP. HOXA9 forms a repressive complex with nuclear matrix-associated protein SAFB to maintain acute myeloid leukemia. Blood. 2023 Apr 6:blood.2022016528. PMCID: PMC10113176
-
Yun H, Narayan N, Vohra S, Giotopoulos G, Mupo A, Madrigal P, Sasca D, Lara-Astiaso D, Horton SJ, Agrawal-Singh S, Meduri E, Basheer F, Marando L, Gozdecka M, Dovey OM, Castillo-Venzor A, Wang X, Gallipoli P, Müller-Tidow C, Osborne CS, Vassiliou GS, Huntly BJP. Mutational synergy during leukemia induction remodels chromatin accessibility, histone modifications and three-dimensional DNA topology to alter gene expression. Nat Genet. 2021 Oct;53(10):1443-1455. PMCID: PMC7611829
- Sasca D, Yun H, Giotopoulos G, Szybinski J, Evan T, Wilson NK, Gerstung M, Gallipoli P, Green AR, Hills R, Russell N, Osborne CS, Papaemmanuil E, Göttgens B, Campbell P, Huntly BJP. Cohesin-dependent regulation of gene expression during differentiation is lost in cohesin-mutated myeloid malignancies. Blood. 2019 Dec 12;134(24):2195-2208. PMCID: PMC7484777
- Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, Pardo M, Yu L, Choudhary JS, Metzakopian E, Iyer V, Yun H, Park N, Varela I, Bautista R, Collord G, Dovey O, Garyfallos DA, De Braekeleer E, Kondo S, Cooper J, Göttgens B, Bullinger L, Northcott PA, Adams D, Vassiliou GS, Huntly BJP. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat Genet. 2018 Jun;50(6):883-894. PMCID: PMC6029661
- Horton SJ, Giotopoulos G, Yun H, Vohra S, Sheppard O, Bashford-Rogers R, Rashid M, Clipson A, Chan WI, Sasca D, Yiangou L, Osaki H, Basheer F, Gallipoli P, Burrows N, Erdem A, Sybirna A, Foerster S, Zhao W, Sustic T, Petrunkina Harrison A, Laurenti E, Okosun J, Hodson D, Wright P, Smith KG, Maxwell P, Fitzgibbon J, Du MQ, Adams DJ, Huntly BJP. Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors. Nature Cell Biology. 2017 Sep;19(9):1093-1104. PMID: 28825697 PMCID:PMC5633079
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, Huthmacher C, Gudgin E, Lugo D, Beinke S, Chapman TD, Roberts EJ, Soden PE, Auger KR, Mirguet O, Doehner K, Delwel R, Burnett AK, Jeffrey P, Drewes G, Lee K, Huntly BJ*, Kouzarides T*. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011 478(7370):529-33 *joint senior authors PMCID:PMC3679520
- Huntly BJP, Shigematzu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, Akashi K, Gilliland DG. MOZ-TIF2, but not BCR-ABL, confers properties of leukaemic stem cells to committed murine haematopoietic progenitors. Cancer Cell. 2004; 6: 586-595 PMID:15607963



