Dr Srinjan Basu
Single-cell and single-molecule imaging approaches in stem cell biology
Email: sb451@cam.ac.uk
Laboratory: Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre
Departmental Affiliation: Department of Physiology, Development and Neuroscience
ORCID record: https://orcid.org/0000-0002-1080-979X
Biography
Srinjan trained in physics, chemistry and biology at the University of Cambridge. During his PhD at Harvard University, he developed Raman-based label-free imaging of nuclear architecture to track chromatin live in dividing cells within mouse skin but also to rapidly identify tumours in human tissue. He also co-authored one of the first papers imaging single transcription factors as they interact with chromatin in live mammalian cells.
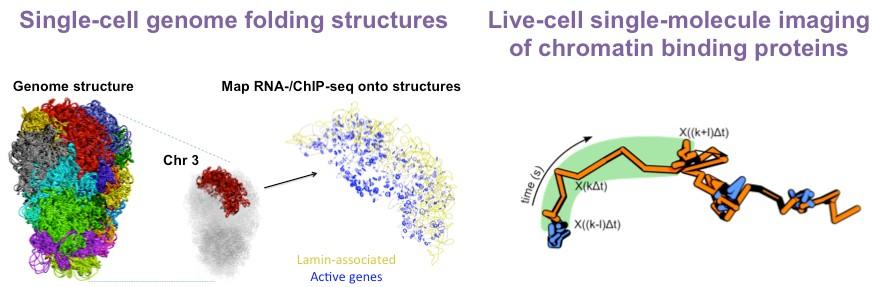
He then returned to Cambridge as a postdoc, where he continued to pioneer approaches that allow the field to probe chromatin at single-molecule and single-cell resolution. He used single-cell Hi-C to determine the architecture of the entire mouse genome at the level of a single pluripotent cell. This revealed considerable cell heterogeneity in the relative positions of enhancers and promoters. Furthermore, it uncovered nuclear hubs enriched for enhancers and promoters but also transcription factors. These studies lay the foundation for and emphasise the importance of probing genome structures at the single-cell level in heterogeneous cell populations during lineage specification.
To further reveal how single proteins and even protein complexes assemble on chromatin, he developed 3D single-molecule tracking and live-cell single-molecule FRET. He has alsoestablished machine-learningalgorithms that classify different types of motion toreveal the chromatin binding kinetics of these key protein complexes but alsoto determine how they control chromatin movement at specific genomic loci.
As a Group Leader at CSCI, he is now using these methodologies to address how chromatin regulators influence mammalian embryo development. He has revealed the chromatin binding kinetics of key chromatin regulators such asheterochromatin protein HP1gamma and a chromatin remodeller called the NuRD complex. He has also recently shown how the NuRD complex modulates enhancer movement to control transcription.
His work increases collaboration between mathematicians, physicists, chemists and biologists and trains a new generation of interdisciplinary researchers. His work increases technology development in bioimaging and next-generation sequencing, training students in interdisciplinary and data-driven biology involving analysis of complex imaging and ‘omics datasets.
Furthermore, as chair of equality, diversity and inclusion, he is pushing his own lab and the wider research community towards better and more inclusive research practices.
Research
The Basu Lab focuses on developing single-cell and single-molecule imaging approaches to improve our understanding of stem cell renewal and differentiation.
In particular, they are interested in how chromatin binding proteins regulate genome architecture and gene expression during stem cell fate transitions and why they are often misregulated during early cancer progression. Single-cell approaches are key to understanding how these proteins work due to the considerable cell heterogeneity that occurs during stem cell fate transitions.
In recent years, the Basu Lab has developed several biophysical and computational approaches to answer these questions. For example, they have established a method combining imaging and single-cell Hi-C to study genome architecture inside individual embryonic stem cells. To understand how proteins interact with each other and with chromatin, they set up several in vitro and live-cell single-molecule imaging approaches capable of localising single proteins at <15 nm resolution.
The Lab is continuing to develop novel single-cell and single-molecule imaging approaches but also using the techniques described above to gain insight into the role of key protein complexes involved in stem cell differentiation.
Group members
(left to right) Srinjan Basu, Oliver Davis, Deep Adhya, Maike Steindel, Stanley Strawbridge, Annabelle Wurmser, Maciej Zurowski, Laura Říhová, Sebastian Berger
Plain English
Stem cells do not have a precise ‘job’ yet, but they can differentiate to become specialized cells, such as the ones that form our bones, our heart or our skin. Our research focuses on how stem cells transition into these different cell types.
In particular, we are interested in what happens to the DNA during these transitions. A molecule of DNA is over two meters long, and yet it can squeeze itself to fit inside the microscopic cells that form our bodies. This packaging process is not done randomly: in fact, a great number of proteins tend to the DNA to fold it into a precise 3D structure that will allow the cell to work properly. These molecules can also reshape the 3D folding in response to what a stem cell needs at any given moment, for example when it goes through differentiation.
When the proteins that fold DNA work incorrectly, it can often result in cancers or neurological disorders. Studying these molecules may therefore help us to develop better drugs or therapies for these conditions.
This is why we have developed methods to visualise these processes. For example, we created an approach that allows us to see for the first time how the entire mammalian genome folds inside a single stem cell. We have also built a microscope that lets us follow individual proteins ‘live’ as they fold the DNA in stem cells. Now we want to use these tools to look at what happens when the cells differentiate, and how folding errors could make them cancerous.
Key Publications
- Steindel M, de Almeida IO, Strawbridge S, Chernova V, Holcman D, Ponjavic A, Srinjan Basu S (2022). Studying the Dynamics of Chromatin-Binding Proteins in Mammalian Cells Using Single-Molecule Localization Microscopy. Chromosome Architecture 209-247
- Basu S*, Needham L-M*, Lando D, Taylor EJR… Eggeling C, Hendrich B, Klenerman D, Lee SF, Laue ED (2018). FRET enhanced photostability allows improved single-molecule tracking of proteins and protein complexes in live mammalian cells. Nat Comm 9(1): 2520. PMCID:PMC6023872 (*equal contribution)
- Stevens TJ*, Lando D*, Basu S*, Atkinson LP… Wutz A, Hendrich B, Klenerman D, Laue ED (2017). 3D structure of individual mammalian genomes studied by single cell Hi-C. Nature 544(7648): 59-64. PMCID:PMC5385134 (*equal contribution)
- Lando D*, Basu S*, Stevens TJ*, Riddell A… Lee SF, Hendrich B, Klenerman D, Laue ED (2017). Combining fluorescence imaging with Hi-C to study 3D genome architecture of the same single cell. Nat Protoc 13(5): 1034. (*equal contribution)
- Fa-Ke Lu*, Basu S*, Igras V, Hoang MP… Fisher DE, Xie XS (2015). Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. PNAS 112 (37): 11624-11629. PMCID:PMC4577158 (*equal contribution)



