Professor Bertie Göttgens
Cellular decision making in normal and leukaemic blood stem cells
Email: bg200@cam.ac.uk
Laboratory: Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre
Departmental Affiliation: Haematology
Biography
Bertie Göttgens currently serves as Director of the Cambridge Stem Cell Institute.
He graduated from Tübingen University in 1992 with a degree in biochemistry. He received his DPhil in biological sciences from the University of Oxford in 1994 and then proceeded to a post-doc position in the Department of Haematology, University of Cambridge, between 1994-2001.
Between 2002-2007 he was a Leukaemia Research Fund Lecturer in the Department of Haematology, Cambridge. He was then a University Lecturer, and subsequently a Reader in Haematology, between 2007-2011.
Since October 2011, Bertie has been Professor of Molecular Haematology, University of Cambridge.
Funding
MRC, Wellcome, Blood Cancer UK, Cancer Research UK, NIH, Aging Biology Foundation
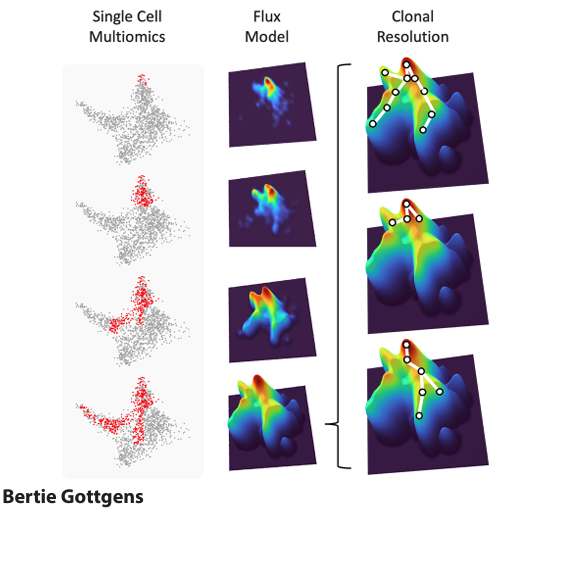
Reconstruction of the haematopoietic differentiation landscape from 45,000 single cell transcriptomes generated from mouse bone marrow (lineage negative / Sca1 positive / Kit positive and lineage negative / Kit positive fractions). Each circle represents a single cell, with the circle colours reporting the results of unsupervised Louvain clustering. The labels indicate the stem/progenitor territory as well as entry points for differentiation trajectories into seven distinct blood lineages.
Research
The Göttgens group uses a combination of experimental and computational approaches to study how blood cells develop, and how mutations in stem/ progenitor cells can cause leukaemia. Over the past 20 years, this integrated approach has delivered several key advancements in the application of genomics to stem cell biology. To begin with, the group illustrated the power of comparative genomics in pinpointing distant gene regulatory elements, followed by the first molecular characterisation of enhancer elements active within haematopoietic stem cells. Subsequent construction of experimentally validated regulatory network models for blood stem and progenitor cells was complemented with comprehensive genome-wide transcription factor maps to provide a holistic view of stem cell regulatory landscapes. Starting in 2010, the group was one of the earliest adopters of new single-cell expression profiling techniques, which they deployed to illuminate stem cell heterogeneity and deliver the first organism-scale single cell maps for gastrulation and early organogenesis. Lastly, the combination of persistent labelling of stem cells with time-series single cell genomics produced the first real-time, quantitative model of in vivo mouse bone marrow haematopoiesis. Current research focuses on (i) early blood development, (ii) dissection of blood stem and progenitor cell differentiation, (iii) functional consequences of leukaemogenic mutations in leukaemia stem/progenitor cells, and (iv) computational modelling of native and perturbed haematopoiesis.
Current research focuses on:
(i) early blood development
(ii) dissection of blood stem and progenitor cell differentiation
(iii) functional consequences of leukaemogenic mutations in leukaemia stem/progenitor cells
(iv) computational modelling of native and perturbed haematopoiesis

Gottgens Group photo
Plain English
Blood stem cells ensure the constant supply of new blood cells throughout a person’s lifetime. The normal function of blood stem cells critically depends on the fine tuning of which genes should be active at any given time. Moreover, a large number of leukaemias arise, when this fine balance of gene activities is disturbed. Through our research, we want to answer the following questions:
• What are the mechanisms that regulate gene activities to ensure normal blood stem cell function?
• Can we identify new strategies to treat leukaemia by reversing the disturbed balance of gene activities?
To answer these questions, we use a combination of experimental and computational approaches. This has allowed us to discover how individual regulatory genes are connected to form complex networks and how perturbation of these complex networks can cause leukaemia.
Key Publications
-
Kucinski, I., Campos, J., Barile, M., Severi, F., Bohin, N., Moreira, P.N., Allen, L., Lawson, H., Haltalli, M.L.R., Kinston, S.J., O’Carroll, D., Kranc, K.R., Göttgens, B. (2023) A time and single-cell resolved model of murine bone marrow hematopoiesis. Cell Stem Cell. 31(2): 244-259
-
Jardine L, Webb S, Goh I, Quiroga Londoño M, Reynolds G, Mather M, Olabi B, Stephenson E, Botting RA, Horsfall D, Engelbert J, Maunder D, Mende N, Murnane C, Dann E, McGrath J, King H, Kucinski I, Queen R, Carey CD, Shrubsole C, Poyner E, Acres M, Jones C, Ness T, Coulthard R, Elliott N, O'Byrne S, Haltalli MLR, Lawrence JE, Lisgo S, Balogh P, Meyer KB, Prigmore E, Ambridge K, Jain MS, Efremova M, Pickard K, Creasey T, Bacardit J, Henderson D, Coxhead J, Filby A, Hussain R, Dixon D, McDonald D, Popescu DM, Kowalczyk MS, Li B, Ashenberg O, Tabaka M, Dionne D, Tickle TL, Slyper M, Rozenblatt-Rosen O, Regev A, Behjati S, Laurenti E, Wilson NK, Roy A, Göttgens B*, Roberts I*, Teichmann SA*, Haniffa M*. Blood and immune development in human fetal bone marrow and Down syndrome. Nature. 2021 Oct;598(7880):327-331. PMCID: PMC7612688
* joint senior author - Stephenson E, Reynolds G, Botting R.A., Calero-Nieto F.J., Morgan M.D., Tuong Z.K., ... Smith K.G.C., Teichmann S.A., Clatworthy M.R., Marioni J.C., Göttgens B*, Haniffa M. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med. 2021 Apr 20. doi: 10.1038/s41591-021-01329-2. PMCID: PMC8121667
* joint senior author
- Pijuan-Sala B., Wilson N.K., Xia J., Hou X., Hannah R.L., Kinston S., Calero-Nieto F.J., Poirion O., Preissl S., Liu F., Göttgens B. (2020). Single-Cell Chromatin Accessibility Maps Reveal Regulatory Programs Driving Early Mouse Organogenesis Nature Cell Biol 22: 487-497 PMCID: PMC7145456
- Kucinski I., Wilson N.K., Hannah R., Kinston S.J., Cauchy P., Lenaerts A., Grosschedl R., Göttgens B. (2020). Cooperation of Lineage-associated Transcription Factors Governs Haematopoietic Progenitor States EMBO Journal 39: e104983 PMCID: PMC7737608
- Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, Mulas C, Ibarra-Soria X, Tyser RCV, Ho DLL, Reik W, Srinivas S, Simons BD, Nichols J, Marioni JC, Göttgens B, (2019) A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566 (7745) 490-495 PMCID: PMC6522369
- Lescroart F, Wang X, Lin X, Swedlund B, Gargouri S, Sànchez-Dànes A, Moignard V, Dubois C, Paulissen C, Kinston S, Göttgens B*, Blanpain C. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science. 2018 Mar 9;359(6380):1177-1181. * joint senior author
- Ibarra-Soria X, Jawaid W, Pijuan-Sala B, Ladopoulos V, Scialdone A, Jörg DJ, Tyser RCV, Calero-Nieto FJ, Mulas C, Nichols J, Vallier L, Srinivas S, Simons BD, Göttgens B*, Marioni JC. Defining murine organogenesis at single-cell resolution reveals a role for the leukotriene pathway in regulating blood progenitor formation. Nat Cell Biol. 2018 Feb;20(2):127-134. * joint senior author
- Scialdone A., Tanaka Y., Jawaid W., Moignard V., Wilson N.K., Macaulay I.C., Marioni J.C., Göttgens B. (2016). Resolving Early Mesoderm Diversification through Single Cell Expression Profiling Nature 535 (7611): 289-29
- Wilson NK, Kent DK, Buettner F, Shehata M, Macaulay IC, Calero-Nieto FJ, Sánchez Castillo M, Oedekoven CA, Diamanti E, Schulte R, Ponting CP, Voet T, Caldas C, Stingl J, Green AR, Theis FJ, Göttgens B. (2015). Combined single cell functional and gene expression analysis resolves heterogeneity within stem cell populations Cell Stem Cell 16(6): 712-724. PMCID:PMC4460190