Professor Tony Green
Haematopoiesis and haematological malignancies
Email: arg1000@cam.ac.uk
Laboratory: Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre
Departmental Affiliation: Haematology
Biography
Tony Green studied medicine (University of Cambridge and University College Hospital, London), subsequently trained in haematology (Royal Free Hospital and Cardiff) and gained his PhD studying oncogenic retroviruses (ICRF, London 1987). Following a post-doctoral period studying haematopoiesis at the Walter and Eliza Hall Institute (Melbourne), he moved to Cambridge in 1991 as a Wellcome Trust Clinical Senior Fellow and Honorary Consultant Haematologist. He was appointed Professor of Haemato-oncology in the University of Cambridge in 1999, served as Head of the University Department of Haematology from 2000-2020, and served as Director of the Wellcome-MRC Cambridge Stem Cell Institute from 2016 until 2022.
His early research explored the transcriptional control of normal blood stem cells and more recently the mechanisms by which such stem cells are subverted to cause haematological malignancies, using the myeloproliferative neoplasms as a tractable model. In work which spans basic, translational and clinical research he identified key causal mutations, described their biological consequences, led practice-changing clinical studies and discovered basic mechanisms of broad relevance for both cancer biology and cytokine signalling.
Tony has held multiple academic, clinical and educational leadership roles, both nationally and internationally, has been appointed to visiting professorships at several universities, was elected Fellow of the Academy of Medical Sciences (2001) and President of the European Haematology Association (2015-2017). Recent awards include the Jean Bernard Award by the European Haematology Association (2020) and the Donald Metcalf award by the International Society for Experimental Hematology (2021).
Funding
Cancer Research UK, WBH Foundation, Alborada Trust
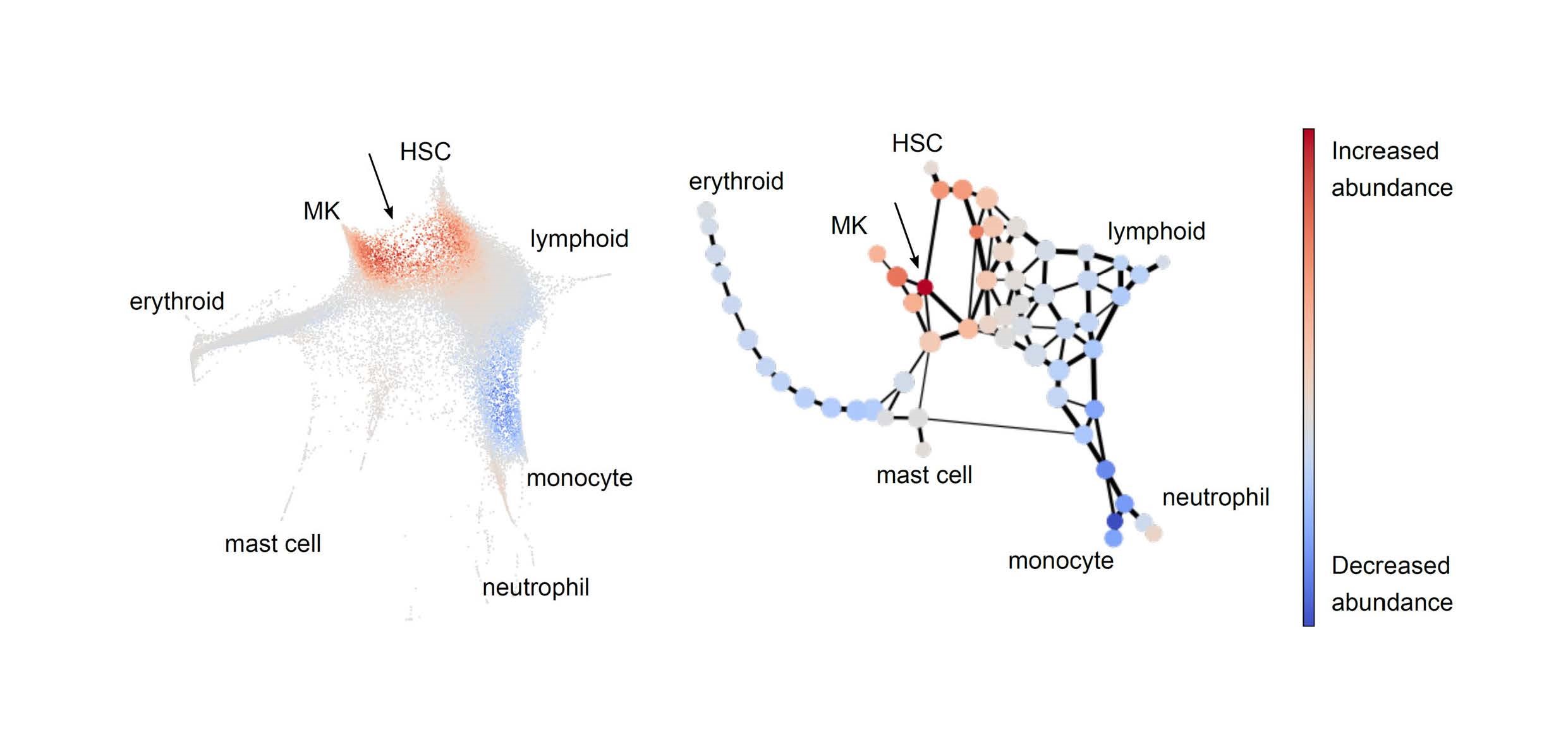
The stem/progenitor landscape is reshaped by mutant CALR in essential thrombocythaemia
Single-cell RNA sequencing of mouse haematopoietic stem and progenitor cells (HSPCs) shows an increase in HSCs and megakaryocyte (MK) progenitor cells in CALR mutant mice, as shown by the arrow in the figure to the left. Force-directed graphical representation of the same data (right) identifies a new intermediate population (arrow) within a trajectory linking HSCs to MK progenitors.
Research
The Green Lab focuses on the mechanisms whereby blood stem cells are subverted during the genesis of haematological malignancies. Over the past two decades they have increasingly concentrated on JAK/STAT signalling which is dysregulated in many cancers and plays a key role in multiple stem cell systems. In particular they have explored the molecular and cellular pathogenesis of a group of pre-leukaemic disorders, the myeloproliferative neoplasms (MPNs), in studies which have spanned basic, translational and clinical research. The myeloproliferative neoplasms harbour mutations that activate the JAK/STAT pathway, are experimentally tractable and provide a paradigm for the earliest stages of tumorigenesis, inaccessible in other cancers.
They described the MPN “mutational landscape” and identified causal mutations which revolutionised their diagnosis and catalysed development of therapeutically valuable JAK-family tyrosine kinase inhibitors. Their more basic research is illuminating the mechanisms whereby the JAK/STAT pathway regulates diverse aspects of cellular function including chromatin biology, DNA replication, genome-wide transcriptional programs and stem cell fate. Recent highlights include: the first demonstration in any cancer that mutation order affects stem and progenitor behaviour, thus influencing clinical presentation, disease outcome and response to therapy; the description of paradigm-shifting non-canonical mechanisms of JAK/STAT signalling; a new biological classification of the MPNs together with personalised predictions based on a knowledge bank; and the use of somatic mutations to reveal human cellular ancestry, stem cell dynamics and life-long clonal evolution.

Green Group photo
Plain English
Blood stem cells produce a large number of cell types and are the best studied adult stem cell system. We are interested in the molecular mechanisms that control the formation and behaviour of normal blood stem cells and how these processes are perturbed during the development of haematological malignancies. In particular we have focused on a group of human pre-leukaemic disorders, called the myeloproliferative neoplasms, in studies which span basic, translational and clinical research. Our basic studies are illuminating the fundamental mechanisms by which these disorders arise, and have a broad relevance for other cancers as well as for the normal process of blood cell formation. Moreover, our results have already had major clinical impact, with new diagnostic approaches embedded in international guidelines, multiple JAK inhibitors in clinical trials and the development of personalised predictions.
Key Publications
- Li J, Williams MJ, Park HJ, Bastos HP, Wang X, Prins D, Wilson NK, Johnson C, Sham K, Wantoch M, Watcham S, Kinston SJ, Pask DC, Hamilton TL, Sneade R, Waller AK, Ghevaert C, Vassiliou GS, Laurenti E, Kent DG, Göttgens B, Green AR. STAT1 is essential for HSC function and maintains MHCIIhi stem cells that resist myeloablation and neoplastic expansion. Blood. 2022 Oct 6;140(14):1592-1606. doi: 10.1182/blood.2021014009. PMID: 35767701.
- Williams N, Lee J, Mitchell E, Moore L, Baxter EJ, Hewinson J, Dawson KJ, Menzies A, Godfrey AL, Green AR*, Campbell PJ*, Nangalia J*. (* Joint senior authors). Life histories of myeloproliferative neoplasms inferred from phylogenies. Nature. 602, 162-168 (2022).
- Prins D, Park HJ, Watcham S, Li J, Vacca M, Bastos HP, Gerbaulet A, Vidal-Puig A, Gottgens B, Green AR. The stem/progenitor landscape is reshaped in a mouse model of essential thrombocythaemia and causes excess megakaryocyte production. Science Advances. 6(48):eabd3139, 2020 (PMID 33239297). PMCID: PMC7688335
- Grinfeld, J+, Nanglia, J+, Baxter EJ, Wedge DC, Angelopoulos N, Cantrill R, Godfrey AL, Papaemmanuil E, Gundem G, MacLean C, Cook J, Mudie L, O’Meara S, Teague JW, Butler AP, Massie CE, Williams N, Nice FL, Anderson CL, Hasselbach HC, Guglielmelli P, McMullin MF, Vannucchi AM, Harrison CN, Gerstung M, Green AR*, Campbell PJ*. (+joint first authors, *joint senior authors). Disease heterogeneity and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 379: 1416-1430, 2018. (PMID 30304655).PMCID: PMC7030948
- Lee-Six H, Øbro NF, Shepherd MS, Grossmann S, Dawson K, Belmonte M, Osborne RJ, Huntly BJP, Martincorena I, Anderson E, O’Neill L, Stratton MR, Laurenti E, Green AR*, Kent DG*, Campbell PJ* (*joint senior authors). Population dynamics of normal human blood inferred from spontaneous somatic mutations. Nature. 561: 473-478, 2018. (PMID 30185910). PMCID: PMC6163040
- Park HJ, Li J, Hnnah R, Biddie S, Leal-Cervantes A, Kirschner K, Flores Santa Cruz D, Sexl V, Gottgens B, Green AR. Cytokine-induced megakaryocytic differentiation is regulated by genome-wide loss of a uSTAT transcriptional program. EMBOJ, doi: 10.15252/embj.201592383. PMID:26702099
- *Ortmann CA, *Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, .... Harrison CN, Vassiliou G, Vannucchi A, Campbell PJ, Green AR. Effect of mutation order on myeloproliferative neoplasms. *Co-first author. N Engl J Med, 372(7):601-12, 2015. PMCID:PMC4660033
- Nangalia J*, Massie CE*, Baxter EJ, Nice FL, Gunes G, Wedge DC…….Papaemmanuil E, Campbell PJ and Green AR. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med, 369(25): 2391-2405, 2013. (*joint first authors). (PMID 24325359)
- Chen E, Beer PA, Godfrey AL, Ortmann CA, Li J, Costa-Pereira AP, Ingle CE, Dermitzakis ET, Campbell PJ, Green AR. Distinct clinical phenotypes associated with JAK2V617F reflect differential STAT1 signaling. Cancer Cell, 18(5): 524-535, 2010. PMCID:PMC2996868
- Dawson MA†, Bannister AJ†, Gottgens B, Foster SD, Bartke T, Green AR*, Kouzarides T* (*joint senior authors; †joint first author). JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature, 461(7265): 819-822, 2009. PMCID:PMC3785147