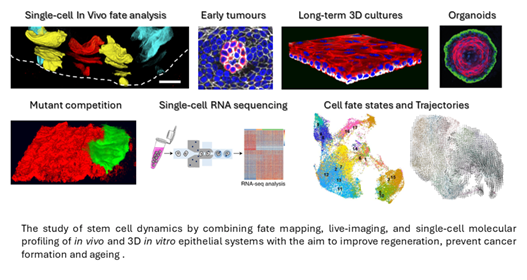
The PhD in Biological Science (Stem Cell Biology) will be carried out under the supervision of a Principal Investigator (PI) from within the Cambridge Stem Cell Institute, and based in their research group.
Please find potential PI projects for 2025-2026 listed below.
Join the Alcolea Group for a project on 'Mapping a blueprint of ageing across tissue scales' (wet lab project).
Keywords: Mechanisms regulating Cell Fate plasticity across epithelial tissues
Project Overview
Despite decades of intense research, modern medicine still faces a critically unresolved question. How can cell fate be manipulated to increase the regenerative capacity of adult tissues? The emerging notion that epithelial cells present a high degree of fate plasticity in response to injury, rewiring their fate in order to preserve tissue integrity, offers the possibility to explore the ill-defined processes modulating cell behaviour in response to regeneration. Here we propose to identify and to characterise the common mechanisms regulating cell plasticity across two different epithelial tissue types, i.e. squamous and the airways epithelium. The ultimate aim of this project is to unearth generic rules dictating the regenerative capacity of epithelial cells. This study will provide a novel perspective on cell fate decision-making, offering a benchmark to develop shared multi-organ regenerative strategies.
This represents a collaborative effort with Dr. Joo-Hyeon Lee based at the Memorial Sloan Kettering, NYC, USA.
Main Techniques
To address this ambitious but critical aspect of epithelial stem cell biology, we will make use of a novel in vivo model that enables tracing the fate of squamous epithelial cells from the earliest stages of commitment towards differentiation. This tool offers a unique opportunity to identify the mechanisms dictating epithelial cell fate plasticity.
We will implement an interdisciplinary approach combining our extensive expertise in in vivo quantitative lineage tracing, modelling cell fate dynamics, single-cell multiomics, mathematical network analysis and 3D organ cultures. Specific techniques will include:
- New long-term 3D Epitheliod cultures (Herms et al. BiorRxiv 2023).
- 3D organoid cultures.
- Clonal lineage tracing.
- Immunostaining of tissue wholemounts.
- 3D whole-organ confocal microscopy.
- Single-cell RNA sequencing and multiomics data mining.
- Gene Regulatory Networks will be assessed using the relevant models, including genetic manipulation via CRISPR/Cas9.
Composite created with images produced by members of the Alcolea Lab and collaborators.
Join the Alcolea Group for a project on 'Impact of ageing on mutant clone competition in squamous epithelial tissues'.
Keywords: Mutant clonal competition, Early tumorigenesis, Squamous epithelia, Epithelial stem cells, Cell fate.
Project Overview
Mounting evidence show that healthy human epithelial tissues accumulate cancer-associated mutations with age, revealing that mutations do not represent the sole cause of cancer. Instead, tumour formation is now believed to depend on a combination of genetic and environmental factors; with competition between adjacent mutant clones acting as critical modulators during early tumorigenesis. However, despite significant efforts in the cancer field, we still know virtually nothing as to how mutant competition is affected by ageing, and whether this contributes to differences in tumour incidence with age.
This PhD project will address the question of how ageing impacts mutant clone dynamics to promote early tumour formation. In particular, we will focus on understanding how mutant clonal competition changes with age, from birth through to late stages in life, using a squamous oesophageal model.
Importantly, work in this area will provide a benchmark to identify new actionable targets of potential use in cancer therapeutics and diagnostics, and to develop strategies to prevent the onset of this aggressive disease.
Main Techniques
To dissect the cellular and molecular mechanisms underlying mutant competition and early tumorigenesis, we will implement a multidisciplinary approach combining our expertise in mutant clonal dynamics in vivo and ex vivo using 3D organ cultures, single-cell multiomics, and mathematical network analysis. Specific techniques will include:
- New long-term 3D Epitheliod cultures (Herms et al. BiorRxiv 2023).
- 3D organoid cultures.
- Clonal lineage tracing.
- Immunostaining of tissue wholemounts.
- 3D whole-organ confocal microscopy.
- Single-cell RNA sequencing and multiomics data mining.
- Gene Regulatory Networks will be assessed using the relevant models, including genetic manipulation via CRISPR/Cas9.
Composite created with images produced by members of the Alcolea Lab and collaborators.
Join the Alcolea Group for a project on 'Mapping a blueprint of ageing across tissue scales' (dry lab project).
Keywords: Ageing, Epithelial stem cells, cell fate plasticity, computational project
Project Overview
The ability of epithelial cells to rewire their programme of cell fate in response to tissue perturbations has emerged as a new paradigm in stem cell biology. This plasticity improves the efficiency of tissue repair by enabling differentiated (non-dividing) cells to reacquire stem/progenitor cell-like behaviour in response to damage. However, despite obvious implications for regenerative medicine, we still know virtually nothing about the processes that regulate promiscuous cell fate programmes, how they impact on tissue physiology, and whether this contributes to age-related complications.
We are seeking candidates with background in Computational Biology to work in a multi-disciplinary PhD project focused on studying epithelial cell fate reprogramming and plasticity during ageing. We are looking for a highly motivated and enthusiastic individual interested in the underlying biological/disease processes and a passion for data driven science. The ultimate aim of this research programme is to identify potential targets to partially reduce/reverse age-related complications.
Main Techniques
In this dry lab project, we will combine our interdisciplinary approach to model epithelial cell fate dynamics during ageing. This will be achieved through single-cell multiomics, gene regulatory network analysis, and quantitative lineage tracing analysis. These will include:
- Analysis of sequencing-based data, particularly single-cell RNA sequencing. Other data may include e.g. WGS, ChIP-seq, ATAC-seq, bulk RNA-seq, multi-omics, and proteomics.
- Downstream data modelling, including pathway analysis, gene regulatory network analysis and cell fate modelling.
- Analysis of integrated multiomics data to decipher regulatory networks associated with tissue ageing. Machine learning approached will enable enumerating and building of possible network topologies between cell types based on cell-cell interaction pairs to study cell communication.
Quantitative lineage tracing analysis using statistical modelling-based approaches. As well as Quantitative image analysis and large-scale image segmentation.
Composite created with images produced by members of the Alcolea Lab and collaborators.
Join the Boroviak Group for a project on 'Functional interrogation of human gastrulation in bioprinted embryo models'.
Keywords: Human embryogenesis, bioprinting, embryo patterning, human development.
Project Overview
How do complex body patterns emerge in the early embryo? The first signs of the human body axis can be traced back to the second week of gestation. To get to this point, the fertilised egg has implanted and established a small sheet of cells, the embryonic disc. Deeply embedded within extraembryonic tissues, gastrulation transforms the EmDisc into three germ layers and organizes the body plan. All of these events are essential for healthy embryo development, but in human they have been notoriously hard to study for ethical and technical reasons.
In mouse, visceral endoderm forms a dynamic signalling centre, the anterior visceral endoderm (AVE), which plays an important role for gastrulation. Soluble inhibitors from the AVE restrict gastrulation towards the opposite side of the embryo, where a combination of WNT-, BMP- and NODAL-signalling induces primitive streak formation. In contrast to the established role of the rodent AVE, the function of the primate AVE in gastrulation is entirely unknown.
Our lab revealed the signalling landscape between implantation and gastrulation of primate embryos in vivo (Bergmann et al., Nature 2022). In this project, we will emulate human gastrulation by printing AVE-like cells onto micropatterned pluripotent stem cells. Primitive streak formation in bioprinted gastruloids will be analysed by 4-colour immunofluorescence confocal image reconstruction and single-cell transcriptome profiling for direct comparison to human and non-human primate embryo gastrula stages. In a second step, we will use knockout and reporter cell lines to pinpoint the individual effects of AVE candidate regulators to devise a conceptional framework for human gastrulation.
Stem-cell-based embryo models elucidating the crosstalk between embryonic and extraembryonic tissues will be critical to understand human implantation failure and how errors in gastrulation can lead to congenital malformations. Ultimately, this research holds the transformative potential to establish patient-specific organogenesis in a dish.
References:
- Bergmann S, Penfold CA, Slatery E, Siriwardena D, Drummer C, Clark S, Strawbridge SE, Kishimoto K, Vickers A, Tewary M, Kohler TN, Hollfelder F, Reik W, Sasaki E, Behr R, Boroviak TE. Spatial profiling of early primate gastrulation in utero. Nature, 2022.
- Connor R. and Boroviak T, Origin and function of the yolk sac in primate embryogenesis, Nature Communications, 2020.
- Munger C, Kohler TN, Slatery E, Ellermann AL, Bergmann S, Penfold CA, Ampartzidis I, Chen Y, Hollfelder F, Boroviak TE. Microgel culture and spatial identity mapping elucidate the signaling requirements for primate epiblast and amnion formation. Development, 2022.
Main Techniques
- Pluripotent stem cell culture
- Extraembryonic stem cell culture
- Stem cell based embryo models
- Single-cell transcriptome and multiomics profiling
- 4-colour confocal imaging and virtual reconstruction
- Micropattern generation and culture
- Bioprinting and microfluidics
- Computational analysis of spatial RNA-seq data in the non-human primate embryos
Fictional representation of our spatial embryo profiling technique. Image credit: Erin Slatery and Thorsten E. Boroviak.
Join the Duque-Correa Group for a project on 'Identifying the key determinants of intestinal epithelia invasion and colonisation by the parasitic worm Trichuris muris'.
Keywords: Intestinal stem cell niche, whipworm, parasitic nematode, infection.
Project Overview
Whipworms infect hundreds of millions of people and cause trichuriasis, a major neglected disease. These large metazoan parasites inhabit a multi-intracellular niche within their host gut lining, where they can remain for years.
Whipworm infection occurs upon ingestion of eggs that hatch in the caecum in a process mediated by the host microbiota. Motile first-stage L1 larvae released from the eggs transverse the mucus layers and enter the intestinal epithelia (IE) at the base of the crypts of Lieberkühn. Our research has shown that whipworm L1 larvae invade intestinal stem, deep secretory and progenitor cells (Duque-Correa et al 2022 Nature Communications); however, the physical and molecular cues directing the larvae and mediating their penetration in the IE are currently unknown. The aim of this PhD project is to identify key determinants of IE invasion and colonisation by whipworms.
This research will provide the first in-depth understanding of whipworms intestinal invasion and colonisation, and guide future studies on the mechanisms underlying larval sensing and tropism, and on invasion pathways with the ultimate prospect of blocking them to prevent infections.
Main Techniques
To address this aim, you will use a novel organoid model developed by the Duque-Correa lab, the first to reproduce whipworm infection of the IE in vitro (Duque-Correa et al 2020 International Journal of Parasitology; Duque-Correa et al 2022 Nature Communications), together with a mouse model of infection with the natural mouse whipworm (Trichuris muris). Using these models, you will characterise interactions between whipworm L1 larvae and the IE that lead to parasite invasion by using transcriptomics and advanced high-resolution imaging techniques (such as confocal, light-sheet and serial block-face SEM), in vitro chemotaxis and biochemical assays.
Join the Duque-Correa Group for a project on 'Studying the impact of parasitic worm infections on intestinal stem cell fate and differentiation'.
Keywords: Stem cell fate, intestinal stem cell niche, whipworm, parasitic nematode, infection.
Project Overview
Whipworms infect hundreds of millions of people and cause trichuriasis, a major neglected disease. These large metazoan parasites inhabit a multi-intracellular niche within their host gut lining, where they can remain for years.
Whipworm infection occurs upon ingestion of eggs that hatch in the caecum in a process mediated by the host microbiota. Motile first-stage L1 larvae released from the eggs transverse the mucus layers and enter the intestinal epithelia (IE) at the base of the crypts of Lieberkühn. Our research has shown that whipworm L1 larvae invade intestinal stem, deep secretory and progenitor cells (Duque-Correa et al 2022 Nature Communications); however, the impact of the parasite on the stem cell fate and stem cell niche remodelling remains unknown. To persist in their host for years, whipworms manipulate the IE renewal cycle (3-5 days) thus avoiding expulsion. Hence, we hypothesise that whipworms induce changes in stem cell division and differentiation, influencing IE architecture and composition.
The aim of this PhD project is to define the effects of whipworm infection on intestinal epithelial stem cell fate and on stromal and immune cell responses that result in changes in mucosal physiology and inflammation, enabling chronic infections.
We anticipate this work will lead to fundamental new knowledge on the whipworm mucosal niche, potentially yielding new targets for anti-parasitic therapies and providing novel insights into how intestinal epithelia repair damage with relevant application toward intestinal inflammatory diseases.
Main Techniques
To address this aim, you will use a novel organoid model developed by the Duque-Correa lab, the first to reproduce whipworm infection of the IE in vitro (Duque-Correa et al 2020 International Journal of Parasitology; Duque-Correa et al 2022 Nature Communications), together with a mouse model of infection with the natural mouse whipworm (Trichuris muris). Using these models, you will characterise gene expression changes (using bulk, single-cell and spatial transcriptomics) of intestinal epithelial cells and stem cell niche populations across infection and after eradication of the parasite. Stem cell remodelling pathways modulated by the parasite will be further studied using lineage tracing and confocal imaging, and stem and stromal cell function will be assessed using organoid assays.
Join the Hodson Group for a project on 'Rethinking the human proteome: bioactive microproteins in non-Hodgkin lymphoma'.
Keywords: Lymphoma, bioinformatics, microproteins
Project Overview
The human genome is believed to contain approximately 20,000 protein-coding genes. These were originally annotated based on criteria for predicting synthesis of stable proteins, including an arbitrary size threshold of 100 amino acids. Technological advances such as Ribosome footprinting (RiboSeq), which reveals the position of every translating ribosome, have abruptly challenged these assumptions, revealing widespread translation of thousands of previously unannotated, non-canonical small open reading frames (smORFs) located in “untranslated” regions or “non-coding” RNAs. The biological impact of this non-canonical translation is beginning to emerge, revealing smORFs that encode microproteins with potent biological functions. However, the vast majority remain completely uncharacterised. This suggests the potential for an entire layer of bioactive molecules that remains almost entirely unexplored.
This collaborative project will leverage a vast collection of RiboSeq data generated within our lab and will combine both computational and advanced wet lab technologies. Our preliminary analysis has already identified thousands of smORFs, many with evidence of stable expression and cell-essential function. We will deploy optimised analytical pipelines that incorporate RiboSeq features and translation factor binding to identify potential microproteins. To provide evidence of functionality, we will perform high throughput base-editor mutagenesis screens, generating a customised, comprehensive gRNA library to mutate the start codon of every predicted smORF in the human translatome. We will perform this start codon mutagenesis screens using cell lines and primary human B cells. These data will be integrated with ultrasensitive, deep proteomic profiling, single cell perturbational transcriptomics and co-expression network analysis to reveal stably expressed microproteins and resolve molecular functions for hundreds of novel, bioactive microproteins. Our mechanistic investigation will focus onto non-Hodgkin lymphoma, where we have the deepest RiboSeq and can leverage the most advanced, already-optimised model systems for functional genomic screening.
Lymphoma is the 6th commonest human cancer and a significant cause of global morbidity and mortality. The slow progress in developing effective new therapies suggests gaps in our understanding. Whilst previous research has predominantly focused on the classical protein-coding genome, we propose that crucial insights into lymphoma biology may lie within the unexplored, non-canonical proteome. Ultimately, our study will provide a comprehensive functional map of non-canonical translation across the human genome, elucidating novel mechanisms that will reshape future research direction in fundamental cell biology and lead to novel therapeutic approaches in lymphoma.
Main Techniques
It is anticipated that a potential student will work as part of a small team of researchers within the Hodson group, and that the student will focus on either the wet lab or the computational aspects of this project, dependent on their prior research experience.
Wet lab techniques will include:
- Genetic manipulation of primary human B cells
- Design and execution of high throughput CRISPR and base editor screens
- Single cell perturbational RNA sequencing.
Computational analysis will include:
- Analysis and integration of large Ribo-Seq, proteomic, genomic and CRISPR/base-editor screen datasets.
- Developing machine learning approaches to define the characteristics of bioactive microproteins.
Join the Khaled Group for a project on 'The tipping point: what drives the progression of breast precancer cells'.
Keywords: Cancer, Precancer, Breast Cancer
Project Overview
Understanding the molecular and cellular mechanisms of how epithelial tissues maintain a homeostatic state throughout the lifespan of mammals is a major challenge for developmental and stem cell biology. From a developmental perspective, the epithelium of the mammary gland is unique as it undergoes most of its development during adulthood. Despite recent efforts of characterising the tissue homeostasis at a cellular level little is known about how this is affected by various developmental processes such as pregnancy or aging and how this might ultimately disrupt epithelial homeostasis resulting in malignant outgrowth. In this study we wish to further our understanding of the changing nature of the differentiation dynamics of the mammary gland. To fully understand how tissue homeostasis is affected by parity and other events it is mandatory to characterise the differentiation dynamics in an age-dependent manner. This becomes evident when looking at epidemiological data. Age is the greatest risk factor for breast carcinomas, and it has been suggested that this is not only due to accumulation of mutations but also due to decreased clonality and selection of clones with proliferative advantage. More importantly, the age-dependent risk of tumorigenesis is modulated by for example parity, which attenuates the risk or predisposing germline mutations which increase the age-associated risk. However, the exact mechanisms and effects of the interaction of these risk factors on tissue homeostasis of the mammary gland remain to be elucidated. Our lab has been using scRNAseq to address these questions by studying the effects of parity, aging and germline mutation (BRCA1/2) on the homeostasis of the mammary gland in mouse and human (PMID:25574598, 29225342, 30127402, 32612221, 33686070, 35091553, 38548988).
For this project the student will be analysing a large dataset of mouse and human scRNAseq which has been collected over several years to identify mechanisms that promote the transition of precancerous cells to cancer. This project will suit someone with some experience in using R and/or Python. In addition, the student will have the opportunity to validate some of the findings using orthogonal spatial technologies with the aim of translating these findings to therapeutic cancer prevention approaches.
Main Techniques
- scRNAseq
- Spatial biology
- Mouse models
Join the Laurenti Group for a project on 'Understanding the stress biology underlying human haematopoietic stem cell adaptation to clinically-relevant ex vivo culture'.
Keywords: Human hematopoietic stem cells, gene therapy, stress biology, single cell biology
Project Overview
Haematopoietic Stem Cell (HSC) Gene Therapy (GT) is the only curative option for many monogenic inherited disorders. It relies on genetic correction of HSCs, the only cells ensuring lifelong blood production when grafted back into the patient. Owing to decades of optimization, current protocols efficiently correct HSC genetic defects. However, they fail to maintain HSC function during the ex vivo culture step, often leading to delayed recovery or graft failure. This functional attrition is a major roadblock in guaranteeing HSC GT safety and outcomes.
Our group recently uncovered that at least 60% of HSC function is lost within the first 24 hours of culture in GT protocol conditions (Johnson et al, Blood 2024), a period of “adaptation” to the culture, during which HSCs mount multiple stress responses. We demonstrate that this loss of HSC function can be dampened 3-fold when HSC are treated with the JAK inhibitor Ruxolitinib during ex vivo culture (Johnson et al, Blood 2024). However, loss of HSC function during adaptation is multifactorial. This PhD project aims to investigate other mechanisms by which HSC function is compromised during culture. It will combine state-of-the-art single cell methods and HSC GT preclinical models to i) identify new stress responses mounted by HSC during adaptation; ii) devise novel methods to minimise ex vivo loss of HSC function and test them in HSC GT preclinical xenograft models.
Globally this project will contribute to our understanding of human HSC biology and identify new pre-clinical strategies to deliver much larger numbers of highly regenerative HSCs to patients.
Main Techniques
- Ex vivo culture of primary human haematopoietic stem and progenitor cells (HSPCs)
- flow cytometry
- single cell transcriptomics and epigenomics
- mRNA overexpression in HSPCs
- gene editing in HSPCs
Blood colonies generated from human haematopoietic stem cells cultured in gene therapy conditions. Image credit: Dr Wenjuan Ma.
Join the Laurenti Group for a project on 'Mechanistic study of how specific mutations associated with clonal haematopoiesis impact human haematopoietic stem cell function'.
Keywords: Human hematopoietic stem cells, clonal hematopoiesis, ageing, single cell biology, gene editing.
Project Overview
Blood cell production is maintained by rare haematopoietic stem cells (HSC). In younger individuals, there are approximately 100,000 HSCs contributing to blood formation. However, with age, HSCs acquire mutations, some of which provide them with a positive clonal selection advantage. Individuals with clonal haematopoiesis, who carry specific mutations over a certain threshold, have increased risk of blood cancers and cardiovascular disease.
Our laboratory recently demonstrated that clonal haematopoiesis is pervasive and that in individuals over the age of 70, less than 20 mutated HSC clones give rise to 1-30% of all blood cells (Mitchell et al., Nature, 2022). In addition, our team and others have identified a large number of mutations that impart a clonal advantage to HSCs. However the cellular and molecular mechanisms by which they do so remain unknown. This PhD project will focus on quantifying the impact of specific mutations (variants) driving clonal expansion on HSC function.
For this, the student will introduce variants via CRISPR gene editing in primary human HSCs then will use tractable state-of-the-art methods to comprehensively assess HSC function, including novel in vitro HSC expansion methods (Sakurai et al., Nature, 2023). This work will be complemented with the study of native mutations in humans. Collectively the project will provide new mechanistic insights into how clonal haematopoiesis related mutations alter blood formation and how that leads to increased disease risk.
Main Techniques
- Ex vivo culture of primary human haematopoietic stem and progenitor cells (HSPCs)
- flow cytometry
- single cell transcriptomics and epigenomics
- mRNA overexpression in HSPCs
- gene editing in HSPCs
{image to follow}
Image credit: Giovanna Mantica. Created with BioRender.com.
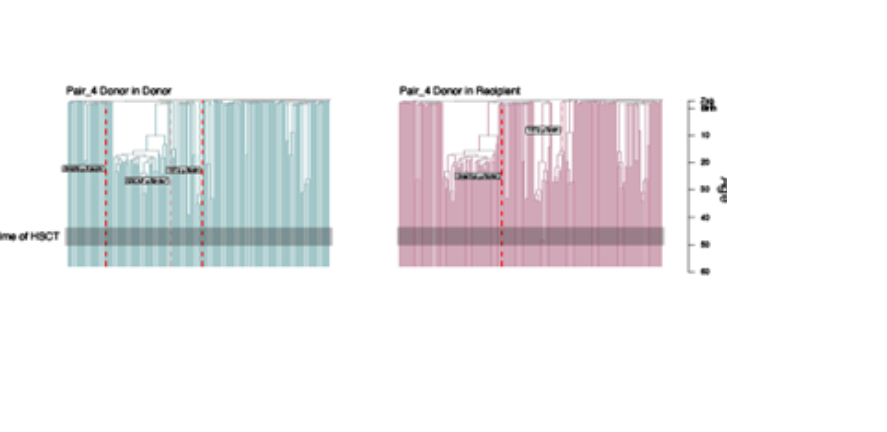
Join the Nangalia Group for a project on 'Investigating intrinsic versus extrinsic haematopoietic ageing through age-mismatched allogeneic stem cell transplantation'.
Project Overview
The blood system declines in function with age with loss of resilience to chemotherapy, decreasing blood counts, defective immunity and increasing risk of blood cancer. Concurrently, haematopoietic clones, some of which harbour cancer-associated driver mutations, gradually expand and start to dominate the haematopoietic system, termed ‘oligo-clonal haematopoiesis’. In addition, there are major changes in the bone marrow micro-environment with a dramatic loss of cellularity, decline in the number and function of mesenchymal stem cells (MSCs), adipocyte accumulation, reduced angiogenesis, and increased secretion of inflammatory molecules.
We don’t know if the emergence of oligo-clonal haemopoiesis, and the associated haematopoietic dysfunction are primarily due to cell-intrinsic (i.e. heritable changes within the haematopoietic cells) or cell-extrinsic (i.e. abnormalities in the microenvironment) mechanisms. Understanding this is a fundamental question that will help inform approaches to prevent the negative effects of haematopoietic ageing. The challenge is that it is rarely possible to ‘decouple’ these two aspects of human ageing.
Age-mismatched allogeneic blood and marrow transplants provide a unique opportunity to study this question. In this procedure, which is commonly used as a treatment for blood cancer or inherited haematological disease, the entire haematopoietic system of the recipient is destroyed and replaced with that of the donor. In some cases, the donor and recipient are a generation apart, with an age mismatch of 25-30 years. The donor may be younger than the recipient (i.e. the microenvironment is older than the haematopoietic cells), while in the other cases the reverse is true (i.e. the microenvironment is younger than the haematopoietic cells).
This project will use whole genome sequencing, methylation profiling, and phylogeny reconstruction of haematopoietic cells from transplant donors and recipients as done in a previous studies (https://www.researchsquare.com/article/rs-2868644/v1 ). This will be used to track and compare donor-derived haematopoietic stem cells from donors and recipients to assess how differing micro-environments affect the clonal dynamics and haematopoietic outputs of the same initial haematopoietic system.
Main Techniques
- Whole genome sequencing
- Whole genome methylation sequencing
- Bio-informatic analyses to reconstruct somatic phylogenies
Image Credit: Comparison of “Donor in donor” and “Donor in recipient” haematopoietic systems, from the donor and recipient of a sibling matched haematopoietic cell transplant (from ‘Clonal dynamics after allogeneic haematopoietic cell transplantation using genome-wide somatic mutations’, https://doi.org/10.21203/rs.3.rs-2868644/v1).
Join the Sampaziotis Group for a project on 'Characterising the circadian clock in human liver'.
Keywords: Circadian clock, machine perfusion, liver.
Project Overview
The circadian clock plays a key role in maintaining homeostasis in all cells. Clock dysregulation desynchronises cell responses to stimuli such as metabolic stress and disrupts the efficacy of these homeostatic mechanisms. Importantly, clock dysregulation is emerging as a key factor in the pathogenesis of a constantly increasing number of human disorders.
Nonetheless, studies in circadian rhythm are limited, as they require serial biopsies over a very short period of time. This is not possible in human, due to the risk of complications; while even in animals, a separate animal needs to be culled for each timepoint which introduces error associated with animal-to-animal variability. To overcome this challenge, we developed a platform, which will allows us to maintain human organs in near physiological conditions outside the body for several days and perform serial biopsies. We propose to use this system to characterise for the first time the circadian clock in human organs. We propose to focus on the liver as a proof-of-principle organ for multiple reasons. Animal data suggest that clock dysregulation plays a key role for metabolic liver disorders, metabolic liver disease is one of the most common causes for liver transplantation and perfused livers can be transplanted back into patients. This gives us for the first time the opportunity to intervene on the clock, by synchronising liver cells ex situ prior to transplantation.
To achieve this goal, we propose to characterise the clock using snRNAseq in healthy human livers perfused ex-situ and validate our results in organoids. We will then perform the same analysis in livers with metabolic disease and use clock synchronisers to explore whether we can improve the function of these organs.
Main Techniques
- Single cell RNA sequencing
- Ex vivo machine perfusion
- Organoid culture
- Basic molecular biology techniques (QPCR, immunofluorescence, western blot)
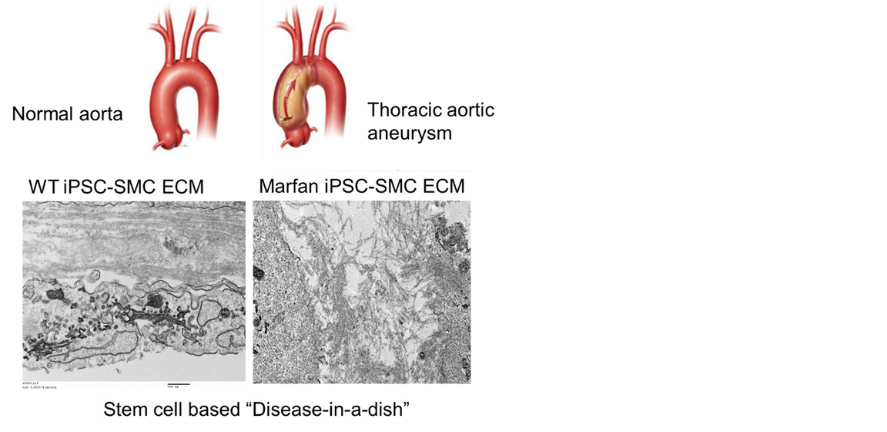
Join the Sinha Group for a project on 'Patient outcomes and new therapies in aortic aneurysm using iPSC ‘disease-in-a-dish’ models'.
Keywords: Aortic aneurysm, iPS cells, disease in a dish, stem cells.
Project Overview
We have pioneered the use of induced pluripotent stem cells from patients with genetically mediated aortic aneurysms (such as Marfan and Loeys-Dietz syndromes) to set up ‘disease-in-a-dish’ in vitro models (PMIDs: 27893734, 36669494). In this project, these systems will be used to define pathological mechanisms, predict patient outcome and response to therapies and to carry out CRISPR-mediated drug target screening. These translational studies will impact patient care and clinical trial design in the future.
Main Techniques
- Stem cell culture and differentiation
- CRISPR genetic correction and screening
- transcriptomics, proteomics
- engineered vascular tissues
- imaging
Image credit: Granata et al. Nature Genetics 2017;49:97-109. PMID: 27893734.
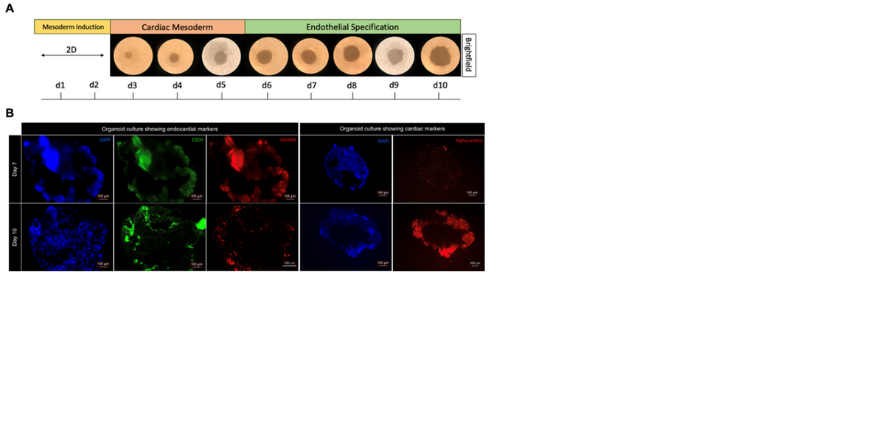
Join the Sinha Group for a project on 'Mechanisms underlying human cardiovascular development'.
Keywords: Cardiovascular development.
Project Overview
We have recently generated a high resolution single cell multiomic atlas of human heart development and used this to establish stem cell derived in vitro models of endocardium and coronary endothelium as well as coronary artery smooth muscle cell development. We have also generated a cardiac organoid that incorporates endocardium and coronary endothelial development. These in vitro systems and organoids will now be further characterised and used to define the molecular mechanisms regulating critical transitions in cardiac development, such as endocardial to endothelial transition or the development of compact vs trabeculated cardiomyocytes.
Main Techniques
This project will provide practical experience in human ESC and iPS cell culture and a variety of cellular/molecular techniques and offer an introduction to the field of regenerative cardiovascular medicine.
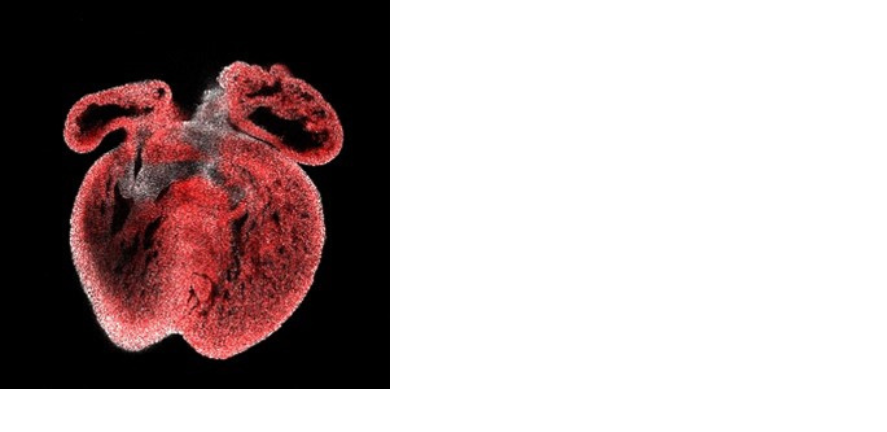
Cardioids generated from human embryonic stem cells (A) showing cardiac and endocardial markers (B).
Data generated by Bea Waller.
Join the Storer Group for a project on 'Unravelling The Parallels Between Regeneration & Tumorigenesis'.
Keywords: Regeneration, Tumour Formation, Cell Biology, Molecular Biology, Genetics.
Project Overview
Some organisms, such as salamanders, retain a remarkable capacity for regeneration in various organs, which occurs through the process of epimorphic regeneration. In mammals, however, multi-tissue regeneration is restricted to the distal digit tip. Here, a blastema forms, a transient mass of proliferating cells, which assemble through activation of stem cells or de-differentiation of mature cells. Interestingly, it has been suggested that the regeneration process may restrain oncogenic transformation. Studies have demonstrated that, in newts, exposure of the dorsal and ventral iris to a carcinogen showed that the dorsal iris, which regenerates, was resistant to transformation. In contrast, the ventral iris, which cannot regenerate was more susceptible to tumour induction. Furthermore, it has been demonstrated that blastema cells are also resistant to tumour formation. Therefore identifying the mechanisms that assures high fidelity of the regeneration process holds great promise for the development of therapeutics that may limit tumorigenesis.
Main Techniques
This project uses the mouse digit tip as a model to identify the signals that limit tumour formation during regeneration. Cutting edge transcriptomics (single cell RNA sequencing and spatial transcriptomics) will be used to identify the key regenerative signalling pathways associated with this process, while genetic models and pharmacological inhibition will be used to
determine their function. The student will also use techniques such as micro CT, confocal microscopy and in vitro culture assays. The successful candidate will join a supportive and hardworking team of scientists based at the Cambridge Stem Cell Institute.
Applications can be made by emailing a one-page CV, full transcript of university results, and half page motivation letter to the academic supervisor listed with the position (Dr Storer, ms2786@cam.ac.uk).
Uninjured mouse digit tip stained for vasculature (yellow) and mesenchymal cells (magenta) taken by Lauren Connolly.
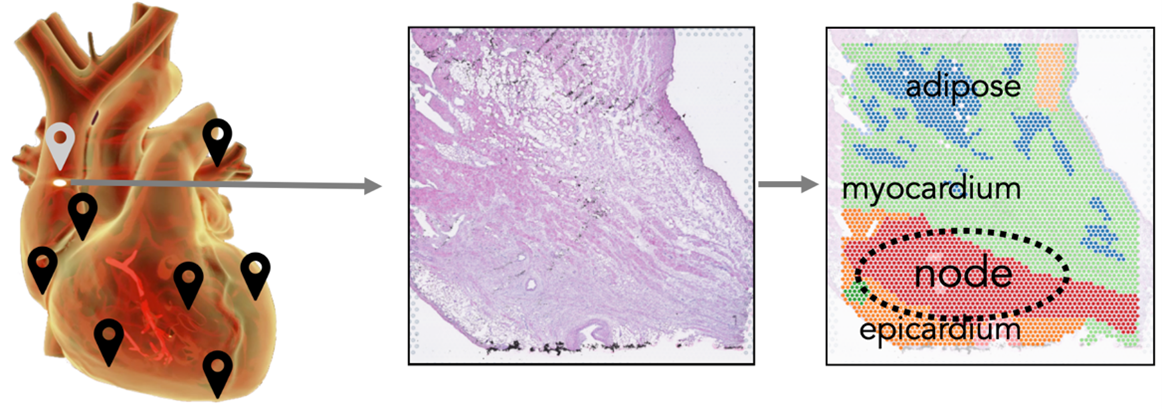
Join the Teichmann Group for a project on 'Cardiac Cell Circuits – deciphering human heart function'.
Keywords: Spatial transcriptomics of the heart.
Project Overview
To define the molecular map of human cardiac tissues, we are applying cell atlas technologies to decipher the molecular mechanism of cell-cell communication in the dozens of tissue niches comprising the human heart. This will contribute to assembling the complete human heart cell atlas, leading to deeper understanding of the function of this organ and its paracrine, endocrine and neural signalling circuits. We apply technologies such as Xenium, VisiumHD and large-volume imaging in an integrative approach to probe novel aspects of heart biology.
We use modelling and machine learning to define a Common Coordinate Framework (CCF) in an Organ-Axis approach (Yayon, Kedlian, Boehme et al, Nature, in press), which could be applied to the lumen-adventitia axis of the big arteries in the heart. Features like cell type abundance or nearest neighbours can then be compared for all three technologies along this axis. Similarly, we will compare tissue niches defined by different technologies and examine if higher resolution technologies can ‘decompose’ the niches with a view to gaining new insights into the cell type composition and cell signalling circuits relevant in heart function.
Main Techniques
Computational analysis of existing data, plus new data generation using cutting edge spatial transcriptomics technology (eg VisiumHD, StereoSeq, Slide-tag).
Image credit: Kazumasa Kanemaru, James Cranley.
Join the Teichmann Group for a project on 'Multiomics to decipher T cell development in the thymus'.
Keywords: T cell development.
Project Overview
Ongoing projects in the Teichmann team are generating scRNA-seq and chromatin accessibility profiles of thymic cells using the 10X multiome kit, combined with multiplex protein staining. The project will examine how altered chromatin states might direct specific T cell fates, examining both T cells and thymic epithelial cells in fetal and paediatric thymi. We have established artificial thymic organoids (ATO) culture systems and will test our insights from single cell data to modulate T cell development in vitro via overexpression of specific transcription factors (TFs) to direct T cell fates. Computational models will be harnessed to predict which TFs may be essential to drive different effector T cell fates.
Main Techiques
- Computational analysis of existing data, plus new data generation using cutting edge TCR repertoire analysis.
- Thymic organoid culture and transfections.
Image credit: Ioannis Sarropoulos.
Join the Teichmann Group for a project on 'Deciphering the human T cell receptor code'.
Keywords: T cell repertoires.
Project Overview
Our laboratory is interested in understanding the establishment of T cell receptor repertoires and the role played by polymorphic peptide-MHC complexes in shaping this. This project will examine recurring motifs in paired alpha and beta chain TCR sequences, and model these with tools such as AlphaFold and utilise deep learning approaches to try to predict binding specificities. Extensive TCR sequencing from single cell data will also from the basis for developing better tools to understand the VDJ rearrangement process, including the identification of D-D, reversed or dual recombination on top of non-functional rearrangements to understand whether these are part of productive TCRs and how these TCRs are distributed within the range of T cell subtypes. This will form the basis for linking TCR motifs to their cognate peptide-MHC targets using data mining of unpublished and public datasets.
Main Techniques
- Computational analysis of existing data, plus new data generation using cutting edge approaches for single cell genomics paired chain TCR repertoire analysis.
- Deep learning and AI tools for structure predictions.
Image credit: Yizhou Yi.
Join the Tyser Group for a project on 'Examining the interplay between form and function during heart development'.
Keywords: Heart Development, Physiology
Project Overview
The heart is the first organ to form and function during development, essential in providing the embryo with sufficient oxygen and nutrients. During heart development, multipotent cardiac progenitors undergo a process of differentiation, whilst also initiating and maintaining contraction. The forming heart is therefore a good model to explore the relationship between function and differentiation/form. Our group is interested in how cardiac function may act as a signalling component to regulate differentiation and heart formation. This work will provide insight into the origins of congenital heart disease and contribute mechanistic understanding as to how the reactivation of developmental gene programs impacts heart disease.
Main Techniques
-
Whole mount imaging
-
Timelapse imaging
-
Single cell RNAseq analysis.
-
Mouse embryology and dissection
-
Transgenic model generation.
Image Credit: Richard Tyser.
Join the Tzelepis Group for a project on 'Cancer-associated sensitisation mechanisms to selective RNA methyltransferase inhibition'.
Keywords: Epitranscriptomics, Novel Therapies, Cancer, RNA Biology.
Project Overview
Epitranscriptomics, the modulation of RNA function via its chemical modification, has emerged as a pervasive new mechanism of gene regulation. The role of the epitranscriptome on stem cell fate and maintenance remains largely unexplored. Similarly, the role of aberrant RNA modification and editing on oncogenic transformation and cancer stem cell maintenance remains poorly investigated, however early studies suggest that the modification of RNA could be exploited for the development of new therapies for devastated diseases including cancer. Our lab has a track record in the discovery and mechanistic characterisation of enzymes that modify RNA as well as the development and characterisation of small molecule inhibitors targeting such enzymes in cancer. Notably, small molecule inhibition of RNA methylation is currently being evaluated in Phase 1 cancer trials, illustrating promising outcomes. Here, we propose to expand this clinically relevant investigations in order to study how particular molecular pathways regulate response to pharmacological inhibition of RNA methylation. We expect the outcomes of this PhD proposal will: (a) lead to currently unknown or incompletely understood fundamental RNA biology, (b) provide a strong mechanistic and functional understanding for the currently unknown roles of selected epitranscriptomic therapies and (c) define new druggable epitranscriptomic combinations in cancer and beyond.
Main Techniques
- CRISPR mutagenesis
- mammalian cell culture
- in vitro and in vivo drug treatment
- molecular cloning
- proteomics
- transcriptomics
- RNA mass spectrometry
Join the Vassiliou Group for a project on 'Therapeutic vulnerabilities of TET2-mutant clonal haematopoiesis'.
Keywords: Clonal haematopoiesis, acute myeloid leukaemia, leukaemia prevention, haematopoietic stem cells, TET2, DNMT3A.
Project Overview
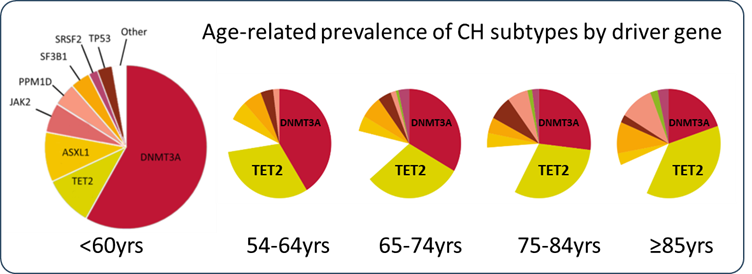
Clonal haematopoiesis (CH), the clonal expansion of a haematopoietic stem cell (HSC) and its progeny driven by somatic mutations, is the shared pre-clinical ancestor of all types of myeloid malignancies (MM), including acute myeloid leukaemia (AML), myelodysplastic syndromes (MDS) and myeloproliferative neoplasms (MPN). CH becomes increasingly common with age to affect >30% of people over 70yrs.
Somatic mutations in the TET2 gene are the second most common cause of CH overall, and the most common cause amongst individuals aged 70 years or older (see figure). CH driven by biallelic mutations in TET2 gene carries a high risk of progression to MM.
This project will investigate the pathogenesis and therapeutic vulnerabilities of TET2-mutant CH using mouse models, primary human cells from volunteers with this from of CH and genetically edited human stem cells cultured in vitro.
Main Techniques
- CRISPR screens
- clonal competition studies
- metabolomics
- investigation of existing and novel therapeutics in vitro and in-vivo (mouse models)
Image credit: G. Vassiliou.
Join the Wilkinson Group for a project on 'Mechanisms regulating blood stem cell self-renewal and differentiation'.
Keywords: Blood stem cell; haematopoietic stem cell; haematopoiesis; cell therapy; gene therapy.
Project Overview
The Wilkinson research group focuses on the biology and translational applications of blood-forming haematopoietic stem cells (HSCs). Self-renewing multipotent HSCs are a rare but incredibly important cell type. Just ~100,000 HSCs generate all the cells of the blood and immune systems, which constitute ~90% of the cells within our bodies. As a long-lived stem cell population, HSCs gradually accumulate genetic mutations and are therefore thought to be a cell-of-origin for several haematological malignancies, particularly myelodysplastic syndrome and myeloid leukaemias.
HSCs are also therapeutically significant with HSC transplantation being the mainstay treatment for the most severe disorders of haematopoiesis ranging from immunodeficiencies to myeloid and lymphoid malignancies. Allogeneic HSC transplantation currently represents the only curative treatment option for a number of haematological malignancies. However, the availability of suitable donor HSCs is a major bottleneck in the use of this therapeutic option. The advent of CRISPR/Cas9 has also led to a renewed interest in HSC gene therapies using autologous HSC transplantation for various genetic diseases.
We have recently developed novel polymer-based culture systems that stably expand HSCs ex vivo, as well as methods to perform efficient genetic manipulation in these cultures. We are now applying these technologies to better understand the biology of healthy and malignant HSCs in order to facilitate the development of new haematological disease treatments.
The PhD project will aim to address biological questions related the mechanisms regulating healthy and malignant HSC activity, and/or aim to develop novel HSC-based therapies. This could include:
- Characterising novel regulators of HSC self-renewal and differentiation;
- Interrogating pre-malignant HSCs and leukaemic transformation;
- Developing novel HSC-based cell and gene therapies.
Key references
- Wilkinson et al., Nature 2019. https://pubmed.ncbi.nlm.nih.gov/31142833/
- Wilkinson et al., Nature Communications 2021. https://pubmed.ncbi.nlm.nih.gov/33514718/
- Igarashi et al., Blood Advances 2022. https://pubmed.ncbi.nlm.nih.gov/36809781/
- Haney et al., bioRxiv 2022. https://www.biorxiv.org/content/10.1101/2022.07.22.501030v1
- Sakurai et al., Nature 2023. https://pubmed.ncbi.nlm.nih.gov/36813966/
Main Techniques
- Primary mouse and/or human haematopoietic stem cell (HSC) isolation and ex vivo culture (expansion, differentiation)
- HSC gene editing (to generate gene knockouts and knockins) and viral transduction
- Multicolour flow cytometry and fluorescence-activated cell sorting
- Clonal barcoding studies
- Omic assays (RNA-seq, ChIP-seq, ATAC-seq)
- Molecular cloning
Join the Zilbauer Group for a project on 'Using human organoids to unravel epigenetic mechanisms in health and Inflammatory Bowel Diseases'.
Keywords: Epigenetics.
Project Overview
The intestinal epithelium provides a critical barrier that contributes towards the establishment of a finely tuned homeostasis in the human gut. Development of a fully functioning intestinal epithelium requires the complex interaction with the environment, mediated through epigenetic mechanisms such as DNA methylation. This project investigates the impact of environmental factors on early epigenetic programming in the human intestinal epithelium and the implications to Inflammatory Bowel Disease pathogenesis.
Main Techniques
The project will utilise human mucosa derived intestinal epithelial organoids (derived from human fetal gut as well as healthy and diseased mucosal biopsies). Organoids will be subjected to gene editing, co-culture and genome wide molecular profiling (including epigenetic profiling and single cell RNA sequencing).
Image credit: Tom Dennison (former PhD student Zilbauer lab).